Conserved allomorphs of MR1 drive the specificity of MR1-restricted TCRs
- PMID: 39445059
- PMCID: PMC11496959
- DOI: 10.3389/fonc.2024.1419528
Conserved allomorphs of MR1 drive the specificity of MR1-restricted TCRs
Abstract
Background: Major histocompatibility complex class-1-related protein (MR1), unlike human leukocyte antigen (HLA) class-1, was until recently considered to be monomorphic. MR1 presents metabolites in the context of host responses to bacterial infection. MR1-restricted TCRs specific to tumor cells have been described, raising interest in their potential therapeutic application for cancer treatment. The diversity of MR1-ligand biology has broadened with the observation that single nucleotide variants (SNVs) exist within MR1 and that allelic variants can impact host immunity.
Methods: The TCR from a MR1-restricted T-cell clone, MC.7.G5, with reported cancer specificity and pan-cancer activity, was cloned and expressed in Jurkat E6.1 TCRαβ- β2M- CD8+ NF-κB:CFP NFAT:eGFP AP-1:mCherry cells or in human donor T cells. Functional activity of 7G5.TCR-T was demonstrated using cytotoxicity assays and by measuring cytokine release after co-culture with cancer cell lines with or without loading of previously described MR1 ligands. MR1 allele sequencing was undertaken after the amplification of the MR1 gene region by PCR. In vivo studies were undertaken at Labcorp Drug Development (Ann Arbor, MI, USA) or Epistem Ltd (Manchester, UK).
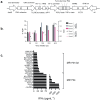
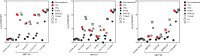
Results: The TCR cloned from MC.7.G5 retained MR1-restricted functional cytotoxicity as 7G5.TCR-T. However, activity was not pan-cancer, as initially reported with the clone MC.7.G5. Recognition was restricted to cells expressing a SNV of MR1 (MR1*04) and was not cancer-specific. 7G5.TCR-T and 7G5-like TCR-T cells reacted to both cancer and healthy cells endogenously expressing MR1*04 SNVs, which encode R9H and H17R substitutions. This allelic specificity could be overcome by expressing supraphysiological levels of the wild-type MR1 (MR1*01) in cell lines.
Conclusions: Healthy individuals harbor T cells reactive to MR1 variants displaying self-ligands expressed in cancer and benign tissues. Described "cancer-specific" MR1-restricted TCRs need further validation, covering conserved allomorphs of MR1. Ligands require identification to ensure targeting MR1 is restricted to those specific to cancer and not normal tissues. For the wider field of immunology and transplant biology, the observation that MR1*04 may behave as an alloantigen warrants further study. .
Keywords: (TCR) T-cell receptor; MR1; T-cell; alloreactive; cancer.
Copyright © 2024 Cornforth, Moyo, Cole, Lam, Lobry, Wolchinsky, Lloyd, Ward, Denham, Masi, Qing Yun, Moore, Dhaouadi, Besra, Veerapen, Illing, Vivian, Raynes, Le Nours, Purcell, Kundu, Silk, Williams, Papa, Rossjohn, Howie and Dukes.
Conflict of interest statement
Authors TC, NM, SC, EL, TL, RW, AL, KW, ED, GM, PQY, CM, SD, SK, JS, LW, SP, DH, JD were employed by Enara Bio Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. JLN declared that they were an editorial board member of Frontiers at the time of submission. This had no impact on the peer review process and the final decision.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

