Exploring the Impact of In Vitro-Transcribed mRNA Impurities on Cellular Responses
- PMID: 39445393
- PMCID: PMC11542617
- DOI: 10.1021/acs.analchem.4c04162
Exploring the Impact of In Vitro-Transcribed mRNA Impurities on Cellular Responses
Abstract
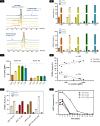
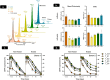
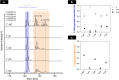
Advances in mRNA technology have enabled mRNA-based therapies to enter a new era of medicine. Such therapies benefit from a single, standardized in vitro transcription (IVT) manufacturing process applicable to a wide range of targets. This process includes several downstream purification steps, which aim to eliminate impurities that potentially affect safety and efficacy. However, it is not fully understood which impurities are the most critical; hence, some efforts are still needed to establish the correlation between RNA impurities and their role in limiting therapeutic efficacy. To study this relationship, we produced in vitro-transcribed mRNAs using several bacteriophage T7 RNA polymerases, including one wild-type and four engineered variants. Important attributes of the mRNA such as integrity, purity, and functional activity were then measured using advanced physicochemical and cellular assays. For impurities including abortive transcripts, mRNAs containing partial poly(A) tails, and double-stranded (ds)RNA byproducts, structure-function relationships have been established by tracking cellular responses (i.e., protein expression, reactogenicity) in multiple cell models. By varying the T7 RNA polymerase, different levels of sense-antisense dsRNA byproducts were measured by mass photometry, contributing directly to immunological reactogenicity in bone marrow-derived dendritic cells. T7 RNA polymerase differences with regard to short (<20 nucleotides) 3'-loopback dsRNA byproducts were also further investigated using native mass spectrometry by precisely resolving these impurities at the nucleotide level. Overall, this study highlights the importance of developing sensitive and advanced analytical methods to characterize IVT mRNA impurities and understand their interaction with cellular machinery in order to ensure quality control of RNA-based therapies.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

