Unraveling the proteomic signatures of coronary artery disease and hypercholesterolemia
- PMID: 39445733
- PMCID: PMC12042678
- DOI: 10.17305/bb.2024.10111
Unraveling the proteomic signatures of coronary artery disease and hypercholesterolemia
Abstract
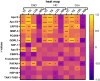
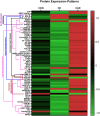
Atherosclerosis, a leading cause of coronary artery disease (CAD), is heavily influenced by hypercholesterolemia (HC). Proteomics research has shown promise in identifying biological markers for CAD diagnosis and prognosis. This cross-sectional study aimed to identify novel biomarkers for detecting HC and CAD. Through the analysis of proteome data from healthy controls (n = 45) and patients diagnosed with HC (n = 51) or CAD (n = 32), distinct protein patterns associated with each condition were identified. Significant alterations in protein levels were identified with a false discovery rate (FDR)-corrected q-value of <0.05. Subsequent receiver operating characteristic (ROC) analysis, with an area under the curve (AUC) greater than 0.75, was conducted. CAD patients exhibited significantly increased levels of the cholesterol-metabolizing protein proprotein convertase subtilisin/kexin type 9 (PCSK9) and varied levels of the angiogenesis-related protein stromal-cell-derived factor-1 (SDF-1) compared to controls. In pairwise comparisons among the study groups, 65 proteins showed significant differential expression. Notably, 14 of these proteins had significant correlations with blood cholesterol levels. Additionally, 22 of the identified proteins were associated with CAD or HC pathways, with nine proteins being common to both conditions (APO E, APO E3, MMP-3, PCSK9, SDF-1, APO B, PAFAH, 60 kDa heat shock protein, and TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 fusion). Nevertheless, this is an exploratory study, and validation studies are needed to confirm these potential protein biomarkers for CAD in the context of HC.
Conflict of interest statement
Conflicts of interest: Authors declare no conflicts of interest.
Figures


References
-
- Tsao CW, Aday AW, Almarzooq ZI, Anderson CAM, Arora P, Avery CL, et al. Heart disease and stroke statistics-2023 update: a report from the american heart association. Circulation. 2023;147(8):e93–e621. https://doi.org/10.1161/CIR.0000000000001137. - PubMed
-
- Jebari-Benslaiman S, Galicia-García U, Larrea-Sebal A, Olaetxea JR, Alloza I, Vandenbroeck K, et al. Pathophysiology of atherosclerosis. Int J Mol Sci. 2022;23(6):3346. https://doi.org/10.3390/ijms23063346. - PMC - PubMed
-
- Prasad K, Mishra M. Mechanism of hypercholesterolemia-induced atherosclerosis. Rev Cardiovasc Med. 2022;23(6):212. https://doi.org/10.31083/j.rcm2306212. - PMC - PubMed
-
- Waldie S, Sebastiani F, Browning K, Maric S, Lind TK, Yepuri N, et al. Lipoprotein ability to exchange and remove lipids from model membranes as a function of fatty acid saturation and presence of cholesterol. Biochim Biophys Acta Mol Cell Biol Lipids. 2020;1865(10):158769. https://doi.org/10.1016/j.bbalip.2020.158769. - PubMed
-
- Tomkin GH. Owens D. LDL as a cause of atherosclerosis. Open Atheroscler Thromb J. 2012;5(1):31–21. https://doi.org/10.2174/1876506801205010013.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

