Genomes of Microtus Rodents Highlight the Importance of Olfactory and Immune Systems in Their Fast Radiation
- PMID: 39445808
- PMCID: PMC11579656
- DOI: 10.1093/gbe/evae233
Genomes of Microtus Rodents Highlight the Importance of Olfactory and Immune Systems in Their Fast Radiation
Abstract
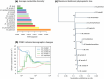
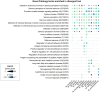
The characterization of genes and biological functions underlying functional diversification and the formation of species is a major goal of evolutionary biology. In this study, we investigated the fast radiation of Microtus voles, one of the most speciose group of mammals, which shows strong genetic divergence despite few readily observable morphological differences. We produced an annotated reference genome for the common vole, Microtus arvalis, and resequenced the genomes of 10 different species and evolutionary lineages spanning the Microtus speciation continuum. Our full-genome sequences illustrate the recent and fast diversification of this group, and we identified genes in highly divergent genomic windows that have likely particular roles in their radiation. We found three biological functions enriched for highly divergent genes in most Microtus species and lineages: olfaction, immunity and metabolism. In particular, olfaction-related genes (mostly olfactory receptors and vomeronasal receptors) are fast evolving in all Microtus species indicating the exceptional importance of the olfactory system in the evolution of these rodents. Of note is e.g. the shared signature among vole species on Olfr1019 which has been associated with fear responses against predator odors in rodents. Our analyses provide a genome-wide basis for the further characterization of the ecological factors and processes of natural and sexual selection that have contributed to the fast radiation of Microtus voles.
Keywords: arvicolinae; genome scan; rapid evolution; reference genome; rodent diversification; voles.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures




References
-
- Baca M, Popović D, Lemanik A, Bañuls-Cardona S, Conard NJ, Cuenca-Bescós G, Desclaux E, Fewlass H, Garcia JT, Hadravova T, et al. Ancient DNA reveals interstadials as a driver of common vole population dynamics during the last glacial period. J Biogeogr. 2023:50(1):183–196. 10.1111/jbi.14521. - DOI

