Persistent tissue-specific resident microbiota in oysters across a broad geographical range
- PMID: 39446070
- PMCID: PMC11500617
- DOI: 10.1111/1758-2229.70026
Persistent tissue-specific resident microbiota in oysters across a broad geographical range
Abstract
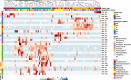
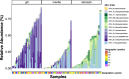
Marine animals often harbour complex microbial communities that influence their physiology. However, strong evidence for resident microbiomes in marine bivalves is lacking, despite their contribution to estuarine habitats and coastal economies. We investigated whether marine bivalves harbour stable, resident microorganisms in specific tissues or if their microbiomes primarily consist of transient members reflecting the environmental microbial pool. Conducting a latitudinal study of wild eastern oysters (Crassostrea virginica) along the East Coast of the United States, we aimed to identify resident microorganisms that persist across a wide geographical range. Our results revealed that microbial communities in seawater and sediment samples followed latitudinal diversity patterns driven by geographic location. In contrast, oyster-associated microbiomes were distinct from their surrounding environments and exhibited tissue-specific compositions. Notably, oyster microbiomes showed greater similarity within the same tissue type across different geographic locations than among different tissue types within the same location. This indicates the presence of tissue-specific resident microbes that persist across large geographical ranges. We identified a persistent set of resident microbiome members for each tissue type, with key microbial members consistent across all locations. These findings underscore the oyster host's role in selecting its microbiome and highlight the importance of tissue-specific microbial communities in understanding bivalve-associated microbiomes.
© 2024 The Author(s). Environmental Microbiology Reports published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Alsterberg, C. , Hulth, S. & Sundbäck, K. (2011) Response of a shallow‐water sediment system to warming. Limnology and Oceanography, 56, 2147–2160. Available from: 10.4319/lo.2011.56.6.2147 - DOI
-
- Andrews, J.D. (1979) Oyster diseases in the Chesapeal Bay. Marine Fisheries Review, 41, 45–53.
-
- Apprill, A. , Mcnally, S. , Parsons, R. & Weber, L. (2015) Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquatic Microbial Ecology, 75, 129–137. Available from: 10.3354/ame01753 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

