Big data research is everyone's research-Making epilepsy data science accessible to the global community: Report of the ILAE big data commission
- PMID: 39446076
- PMCID: PMC11651381
- DOI: 10.1002/epd2.20288
Big data research is everyone's research-Making epilepsy data science accessible to the global community: Report of the ILAE big data commission
Abstract
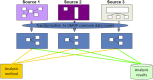
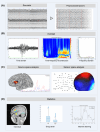
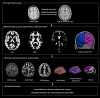
Epilepsy care generates multiple sources of high-dimensional data, including clinical, imaging, electroencephalographic, genomic, and neuropsychological information, that are collected routinely to establish the diagnosis and guide management. Thanks to high-performance computing, sophisticated graphics processing units, and advanced analytics, we are now on the cusp of being able to use these data to significantly improve individualized care for people with epilepsy. Despite this, many clinicians, health care providers, and people with epilepsy are apprehensive about implementing Big Data and accompanying technologies such as artificial intelligence (AI). Practical, ethical, privacy, and climate issues represent real and enduring concerns that have yet to be completely resolved. Similarly, Big Data and AI-related biases have the potential to exacerbate local and global disparities. These are highly germane concerns to the field of epilepsy, given its high burden in developing nations and areas of socioeconomic deprivation. This educational paper from the International League Against Epilepsy's (ILAE) Big Data Commission aims to help clinicians caring for people with epilepsy become familiar with how Big Data is collected and processed, how they are applied to studies using AI, and outline the immense potential positive impact Big Data can have on diagnosis and management.
Keywords: artificial intelligence; big data; common data models; epilepsy; ethics.
© 2024 The Author(s). Epileptic Disorders published by Wiley Periodicals LLC on behalf of International League Against Epilepsy.
Figures

References
-
- Mehta N, Pandit A. Concurrence of big data analytics and healthcare: a systematic review. Int J Med Inform. 2018;114:57–65. - PubMed
-
- Bohr A, Memarzadeh K. The rise of artificial intelligence in healthcare applications. In: Bohr A, Memarzadeh K, editors. Artificial intelligence in healthcare. London, UK: Academic Press is an imprint of Elsevier; 2020. p. 25–60.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

