Genomic Surveillance for SARS-CoV-2 Variants: Circulation of Omicron XBB and JN.1 Lineages - United States, May 2023-September 2024
- PMID: 39446667
- PMCID: PMC11500842
- DOI: 10.15585/mmwr.mm7342a1
Genomic Surveillance for SARS-CoV-2 Variants: Circulation of Omicron XBB and JN.1 Lineages - United States, May 2023-September 2024
Abstract
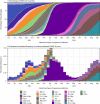
CDC continues to track the evolution of SARS-CoV-2, including the Omicron variant and its descendants, using national genomic surveillance. This report summarizes U.S. trends in variant proportion estimates during May 2023-September 2024, a period when SARS-CoV-2 lineages primarily comprised descendants of Omicron variants XBB and JN.1. During summer and fall 2023, multiple descendants of XBB with immune escape substitutions emerged and reached >10% prevalence, including EG.5-like lineages by June 24, FL.1.5.1-like lineages by August 5, HV.1 lineage by September 30, and HK.3-like lineages by November 11. In winter 2023, the JN.1 variant emerged in the United States and rapidly attained predominance nationwide, representing a substantial genetic shift (>30 spike protein amino acid differences) from XBB lineages. Descendants of JN.1 subsequently circulated and reached >10% prevalence, including KQ.1-like and KP.2-like lineages by April 13, KP.3 and LB.1-like lineages by May 25, and KP.3.1.1 by July 20. Surges in COVID-19 cases occurred in winter 2024 during the shift to JN.1 predominance, as well as in summer 2023 and 2024 during circulation of multiple XBB and JN.1 descendants, respectively. The ongoing evolution of the Omicron variant highlights the importance of continued genomic surveillance to guide medical countermeasure development, including the selection of antigens for updated COVID-19 vaccines.
Conflict of interest statement
All authors have completed and submitted the International Committee of Medical Journal Editors form for disclosure of potential conflicts of interest. No potential conflicts of interest were disclosed.
Figures
References
-
- DeCuir J, Payne AB, Self WH, et al. ; CDC COVID-19 Vaccine Effectiveness Collaborators. Interim effectiveness of updated 2023–2024 (monovalent XBB.1.5) COVID-19 vaccines against COVID-19–associated emergency department and urgent care encounters and hospitalization among immunocompetent adults aged ≥18 years—VISION and IVY networks, September 2023–January 2024. MMWR Morb Mortal Wkly Rep 2024;73:180–8. 10.15585/mmwr.mm7308a5 - DOI - PMC - PubMed
-
- Ma KC, Surie D, Lauring AS, et al. Effectiveness of updated 2023–2024 (monovalent XBB.1.5) COVID-19 vaccination against SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineage hospitalization and a comparison of clinical severity—IVY network, 26 hospitals, October 18, 2023–March 9, 2024. Clin Infect Dis 2024; Epub August 6, 2024. 10.1093/cid/ciae405 - DOI - PubMed
-
- Lambrou AS, Shirk P, Steele MK, et al. ; Strain Surveillance and Emerging Variants Bioinformatic Working Group; Strain Surveillance and Emerging Variants NS3 Working Group. Genomic surveillance for SARS-CoV-2 variants: predominance of the Delta (B.1.617.2) and Omicron (B.1.1.529) variants—United States, June 2021–January 2022. MMWR Morb Mortal Wkly Rep 2022;71:206–11. 10.15585/mmwr.mm7106a4 - DOI - PMC - PubMed
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

