Epi-SSA: A novel epistasis detection method based on a multi-objective sparrow search algorithm
- PMID: 39446852
- PMCID: PMC11500897
- DOI: 10.1371/journal.pone.0311223
Epi-SSA: A novel epistasis detection method based on a multi-objective sparrow search algorithm
Abstract
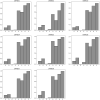
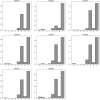
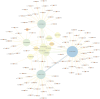
Genome-wide association studies typically considers epistatic interactions as a crucial factor in exploring complex diseases. However, the current methods primarily concentrate on the detection of two-order epistatic interactions, with flaws in accuracy. In this work, we introduce a novel method called Epi-SSA, which can be better utilized to detect high-order epistatic interactions. Epi-SSA draws inspiration from the sparrow search algorithm and optimizes the population based on multiple objective functions in each iteration, in order to be able to more precisely identify epistatic interactions. To evaluate its performance, we conducted a comprehensive comparison between Epi-SSA and seven other methods using five simulation datasets: DME 100, DNME 100, DME 1000, DNME 1000 and DNME3 100. The DME 100 dataset encompasses eight second-order epistasis disease models with marginal effects, each comprising 100 simulated data instances, featuring 100 SNPs per instance, alongside 800 case and 800 control samples. The DNME 100 encompasses eight second-order epistasis disease models without marginal effects and retains other properties consistent with DME 100. Experiments on the DME 100 and DNME 100 datasets were designed to evaluate the algorithms' capacity to detect epistasis across varying disease models. The DME 1000 and DNME 1000 datasets extend the complexity with 1000 SNPs per simulated data instance, while retaining other properties consistent with DME 100 and DNME 100. These experiments aimed to gauge the algorithms' adaptability in detecting epistasis as the number of SNPs in the data increases. The DNME3 100 dataset introduces a higher level of complexity with six third-order epistasis disease models, otherwise paralleling the structure of DNME 100, serving to test the algorithms' proficiency in identifying higher-order epistasis. The highest average F-measures achieved by the seven other existing methods on the five datasets are 0.86, 0.86, 0.41, 0.56, and 0.79 respectively, while the average F-measures of Epi-SSA on the five datasets are 0.92, 0.97, 0.79, 0.86, and 0.97 respectively. The experimental results demonstrate that the Epi-SSA algorithm outperforms other methods in a variety of epistasis detection tasks. As the number of SNPs in the data set increases and the order of epistasis rises, the advantages of the Epi-SSA algorithm become increasingly pronounced. In addition, we applied Epi-SSA to the analysis of the WTCCC dataset, uncovering numerous genes and gene pairs that might play a significant role in the pathogenesis of seven complex diseases. It is worthy of note that some of these genes have been relatedly reported in the Comparative Toxicogenomics Database (CTD). Epi-SSA is a potent tool for detecting epistatic interactions, which aids us in further comprehending the pathogenesis of common and complex diseases. The source code of Epi-SSA can be obtained at https://osf.io/6sqwj/.
Copyright: © 2024 Sun et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
NO authors have competing interests.
Figures






References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

