Evolution of the pheV-tRNA integrated genomic island in Escherichia coli
- PMID: 39446883
- PMCID: PMC11537424
- DOI: 10.1371/journal.pgen.1011459
Evolution of the pheV-tRNA integrated genomic island in Escherichia coli
Abstract
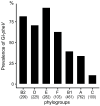
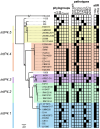
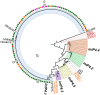
Escherichia coli exhibit extensive genetic diversity at the genome level, particularly within their accessory genome. The tRNA integrated genomic islands (GIs), a part of the E. coli accessory genome, play an important role in pathogenicity. However, studies examining the evolution of GIs have been challenging due to their large size, considerable gene content variation and fragmented assembly in draft genomes. Here we examined the evolution of the GI integrated at pheV-tRNA (GI-pheV), with a primary focus on uropathogenic E. coli (UPEC) and the globally disseminated multidrug resistant ST131 clone. We show the gene content of GI-pheV is highly diverse and arranged in a modular configuration, with the P4 integrase encoding gene intP4 the only conserved gene. Despite this diversity, the GI-pheV gene content displayed conserved features among strains from the same pathotype. In ST131, GI-pheV corresponding to the reference strain EC958 (EC958_GI-pheV) was found in ~90% of strains. Phylogenetic analyses suggested that GI-pheV in ST131 has evolved together with the core genome, with the loss/gain of specific modules (or the entire GI) linked to strain specific events. Overall, we show GI-pheV exhibits a dynamic evolutionary pathway, in which modules and genes have evolved through multiple events including insertions, deletions and recombination.
Copyright: © 2024 Nhu et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

