Cryo-EM structure of a natural prion: chronic wasting disease fibrils from deer
- PMID: 39448454
- PMCID: PMC11502585
- DOI: 10.1007/s00401-024-02813-y
Cryo-EM structure of a natural prion: chronic wasting disease fibrils from deer
Abstract
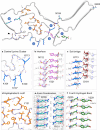
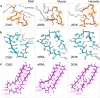
Chronic wasting disease (CWD) is a widely distributed prion disease of cervids with implications for wildlife conservation and also for human and livestock health. The structures of infectious prions that cause CWD and other natural prion diseases of mammalian hosts have been poorly understood. Here we report a 2.8 Å resolution cryogenic electron microscopy-based structure of CWD prion fibrils from the brain of a naturally infected white-tailed deer expressing the most common wild-type PrP sequence. Like recently solved rodent-adapted scrapie prion fibrils, our atomic model of CWD fibrils contains single stacks of PrP molecules forming parallel in-register intermolecular β-sheets and intervening loops comprising major N- and C-terminal lobes within the fibril cross-section. However, CWD fibrils from a natural cervid host differ markedly from the rodent structures in many other features, including a ~ 180° twist in the relative orientation of the lobes. This CWD structure suggests mechanisms underlying the apparent CWD transmission barrier to humans and should facilitate more rational approaches to the development of CWD vaccines and therapeutics.
Keywords: Amyloid; Chronic wasting disease; Cryo-electron microscopy; Deer; Prion; Structure.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Baron GS, Hughson AG, Raymond GJ, Offerdahl DK, Barton KA, Raymond LD et al (2011) Effect of glycans and the glycophosphatidylinositol anchor on strain dependent conformations of scrapie prion protein: improved purifications and infrared spectra. Biochemistry 50:4479–4490. 10.1021/bi2003907 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

