Vaccine-induced human monoclonal antibodies to PfRH5 show broadly neutralizing activity against P. falciparum clinical isolates
- PMID: 39448626
- PMCID: PMC11502735
- DOI: 10.1038/s41541-024-00986-x
Vaccine-induced human monoclonal antibodies to PfRH5 show broadly neutralizing activity against P. falciparum clinical isolates
Abstract
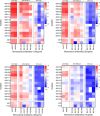
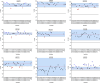
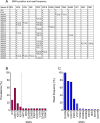
Vaccines to the Plasmodium falciparum reticulocyte binding-like protein homologue 5 (PfRH5) target the blood-stage of the parasite life cycle. PfRH5 has the potential to trigger the production of strain-transcendent antibodies and has proven its efficacy both in pre-clinical and early clinical studies. Vaccine-induced monoclonal antibodies (mAbs) to PfRH5 showed promising outcomes against cultured P. falciparum laboratory strains from distinct geographic areas. Here, we assessed the functional impact of vaccine-induced anti-PfRH5 mAbs on more genetically diverse P. falciparum clinical isolates. We used mAbs previously isolated from single B cells of UK adult PfRH5 vaccinees and used ex-vivo growth inhibition activity (GIA) assays to assess their efficacy against P. falciparum clinical isolates. Next-generation sequencing (NGS) was used to assess the breadth of genetic diversity in P. falciparum clinical isolates and to infer the genotype/phenotype relationship involved in antibody susceptibility. We showed a dose-dependent inhibition of clinical isolates with three main GIA groups: high, medium and low. Except for one isolate, our data show no significant differences in the mAb GIA profile between P. falciparum clinical isolates and the 3D7 reference strain, which harbors the vaccine allele. We also observed an additive relationship for mAb combinations, whereby the combination of GIA-low and GIA-medium antibodies resulted in increased GIA, having important implications for the contribution of specific clones within polyclonal IgG responses. While our NGS analysis showed the occurrence of novel mutations in the pfrh5 gene, these mutations were predicted to have little or no functional impact on the antigen structure or recognition by known mAbs. Our present findings complement earlier reports on the strain transcendent potential of anti-PfRH5 mAbs and constitute, to our knowledge, the first report on the susceptibility of P. falciparum clinical isolates from natural infections to vaccine-induced human mAbs to PfRH5.
© 2024. The Author(s).
Conflict of interest statement
KMc and SJD are inventors on patent applications relating to RH5 malaria vaccines and/or antibodies. All other authors have declared that no conflict of interest exists.
Figures



References
-
- Organization, W. H. World malaria report 2023. (2023).
-
- WHO, U. and. Reversing the Incidence of Malaria 2000–2015. WHO Glob. Malar. Program (2015).
-
- Balikagala, B. et al. Evidence of Artemisinin-Resistant Malaria in Africa. N. Engl. J. Med. 10.1056/nejmoa2101746 (2021). - PubMed
Grants and funding
- UL1 TR001863/TR/NCATS NIH HHS/United States
- K01 TW010496/TW/FIC NIH HHS/United States
- R01 AI168238/AI/NIAID NIH HHS/United States
- R61 AI176583-01/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- 1S10OD030363-01A1/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
LinkOut - more resources
Full Text Sources
Other Literature Sources

