Epigenetic modulation via the C-terminal tail of H2A.Z
- PMID: 39448645
- PMCID: PMC11502880
- DOI: 10.1038/s41467-024-53514-9
Epigenetic modulation via the C-terminal tail of H2A.Z
Abstract
H2A.Z-nucleosomes are present in both euchromatin and heterochromatin and it has proven difficult to interpret their disparate roles in the context of their stability features. Using an in situ assay of nucleosome stability and DT40 cells expressing engineered forms of the histone variant we show that native H2A.Z, but not C-terminally truncated H2A.Z (H2A.Z∆C), is released from nucleosomes of peripheral heterochromatin at unusually high salt concentrations. H2A.Z and H3K9me3 landscapes are reorganized in H2A.Z∆C-nuclei and overall sensitivity of chromatin to nucleases is increased. These tail-dependent differences are recapitulated upon treatment of HeLa nuclei with the H2A.Z-tail-peptide (C9), with MNase sensitivity being increased genome-wide. Fluorescence correlation spectroscopy revealed C9 binding to reconstituted nucleosomes. When introduced into live cells, C9 elicited chromatin reorganization, overall nucleosome destabilization and changes in gene expression. Thus, H2A.Z-nucleosomes influence global chromatin architecture in a tail-dependent manner, what can be modulated by introducing the tail-peptide into live cells.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

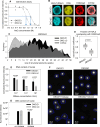

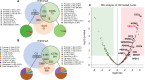
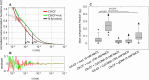


References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

