Improving docking and virtual screening performance using AlphaFold2 multi-state modeling for kinases
- PMID: 39448664
- PMCID: PMC11502823
- DOI: 10.1038/s41598-024-75400-6
Improving docking and virtual screening performance using AlphaFold2 multi-state modeling for kinases
Abstract
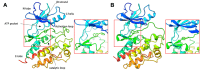
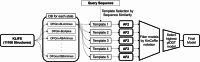
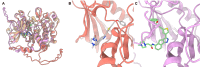
Structure-based virtual screening (SBVS) is a crucial computational approach in drug discovery, but its performance is sensitive to structural variations. Kinases, which are major drug targets, exemplify this challenge due to active site conformational changes caused by different inhibitor types. Most experimentally determined kinase structures have the DFGin state, potentially biasing SBVS towards type I inhibitors and limiting the discovery of diverse scaffolds. We introduce a multi-state modeling (MSM) protocol for AlphaFold2 (AF2) kinase structures using state-specific templates to address these challenges. Our comprehensive benchmarks evaluate predicted model qualities, binding pose prediction accuracy, and hit compound identification through ensemble SBVS. Results demonstrate that MSM models exhibit comparable or improved structural accuracy compared to standard AF2 models, enhancing pose prediction accuracy and effectively capturing kinase-ligand interactions. In virtual screening experiments, our MSM approach consistently outperforms standard AF2 and AF3 modeling, particularly in identifying diverse hit compounds. This study highlights the potential of MSM in broadening kinase inhibitor discovery by facilitating the identification of chemically diverse inhibitors, offering a promising solution to the structural bias problem in kinase-targeted drug discovery.
Keywords: AlphaFold2; Ensemble screening; Kinase; Multi-state modeling; Protein-ligand docking; Structure-based virtual screening.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

