DNA-protein quasi-mapping for rapid differential gene expression analysis in non-model organisms
- PMID: 39448913
- PMCID: PMC11515663
- DOI: 10.1186/s12859-024-05924-1
DNA-protein quasi-mapping for rapid differential gene expression analysis in non-model organisms
Abstract
Background: Conventional differential gene expression analysis pipelines for non-model organisms require computationally expensive transcriptome assembly. We recently proposed an alternative strategy of directly aligning RNA-seq reads to a protein database, and demonstrated drastic improvements in speed, memory usage, and accuracy in identifying differentially expressed genes.
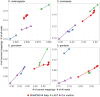
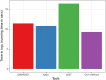
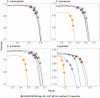
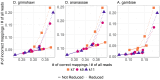
Result: Here we report a further speed-up by replacing DNA-protein alignment by quasi-mapping, making our pipeline > 1000× faster than assembly-based approach, and still more accurate. We also compare quasi-mapping to other mapping techniques, and show that it is faster but at the cost of sensitivity.
Conclusion: We provide a quick-and-dirty differential gene expression analysis pipeline for non-model organisms without a reference transcriptome, which directly quasi-maps RNA-seq reads to a reference protein database, avoiding computationally expensive transcriptome assembly.
Keywords: DNA-protein alignment; Differential gene expression analysis; Non-model organism; Quasi-mapping; RNA-seq.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests
Figures


References
-
- Vijay N, Poelstra JW, Künstner A, Wolf JBW. Challenges and strategies in transcriptome assembly and differential gene expression quantification a. comprehensive in-silico assessment of RNA-seq experiments. Mol Ecol. 2012;22(3):620–34. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

