MYB transcription factors, their regulation and interactions with non-coding RNAs during drought stress in Brassica juncea
- PMID: 39448923
- PMCID: PMC11515528
- DOI: 10.1186/s12870-024-05736-8
MYB transcription factors, their regulation and interactions with non-coding RNAs during drought stress in Brassica juncea
Abstract
Background: Brassica juncea (L.) Czern is an important oilseed crop affected by various abiotic stresses like drought, heat, and salt. These stresses have detrimental effects on the crop's overall growth, development and yield. Various Transcription factors (TFs) are involved in regulation of plant stress response by modulating expression of stress-responsive genes. The myeloblastosis (MYB) TFs is one of the largest families of TFs associated with various developmental and biological processes such as plant growth, secondary metabolism, stress response etc. However, MYB TFs and their regulation by non-coding RNAs (ncRNAs) in response to stress have not been studied in B. juncea. Thus, we performed a detailed study on the MYB TF family and their interactions with miRNAs and Long non coding RNAs.
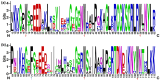
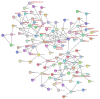
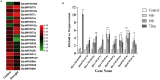
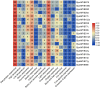
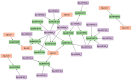
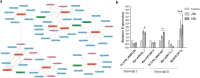
Results: Computational investigation of genome and proteome data presented a comprehensive picture of the MYB genes and their protein architecture, including intron-exon organisation, conserved motif analysis, R2R3 MYB DNA-binding domains analysis, sub-cellular localization, protein-protein interaction and chromosomal locations. Phylogenetically, BjuMYBs were further classified into different subclades on the basis of topology and classification in Arabidopsis. A total of 751 MYBs were identified in B. juncea corresponding to 297 1R-BjuMYBs, 440 R2R3-BjuMYBs, 12 3R-BjuMYBs, and 2 4R-BjuMYBs types. We validated the transcriptional profiles of nine selected BjuMYBs under drought stress through RT-qPCR. Promoter analysis indicated the presence of drought-responsive cis-regulatory components. Additionally, the miRNA-MYB TF interactions was also studied, and most of the microRNAs (miRNAs) that target BjuMYBs were involved in abiotic stress response and developmental processes. Regulatory network analysis and expression patterns of lncRNA-miRNA-MYB indicated that selected long non-coding RNAs (lncRNAs) acted as strong endogenous target mimics (eTMs) of the miRNAs regulated expression of BjuMYBs under drought stress.
Conclusions: The present study has established preliminary groundwork of MYB TFs and their interaction with ncRNAs in B. juncea and it will help in developing drought- tolerant Brassica crops.
Keywords: Brassica juncea; Cis-regulatory elements; MYB domain; Abiotic stress; Expression profile; lncRNAs; miRNA targets.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
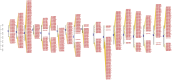
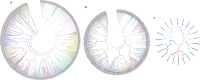
Similar articles
-
Identification, genomic localization, and functional validation of salt-stress-related lncRNAs in Indian Mustard (Brassica juncea L.).BMC Genomics. 2024 Nov 20;25(1):1121. doi: 10.1186/s12864-024-10964-1. BMC Genomics. 2024. PMID: 39567864 Free PMC article.
-
Global insights into high temperature and drought stress regulated genes by RNA-Seq in economically important oilseed crop Brassica juncea.BMC Plant Biol. 2015 Jan 21;15:9. doi: 10.1186/s12870-014-0405-1. BMC Plant Biol. 2015. PMID: 25604693 Free PMC article.
-
Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa.BMC Genomics. 2019 Mar 19;20(1):227. doi: 10.1186/s12864-019-5593-5. BMC Genomics. 2019. PMID: 30890148 Free PMC article.
-
Non-Coding RNAs in Legumes: Their Emerging Roles in Regulating Biotic/Abiotic Stress Responses and Plant Growth and Development.Cells. 2021 Jul 2;10(7):1674. doi: 10.3390/cells10071674. Cells. 2021. PMID: 34359842 Free PMC article. Review.
-
Non-Coding RNAs in Response to Drought Stress.Int J Mol Sci. 2021 Nov 20;22(22):12519. doi: 10.3390/ijms222212519. Int J Mol Sci. 2021. PMID: 34830399 Free PMC article. Review.
Cited by
-
Transcriptomics and network pharmacology analysis reveal key genes in alkaloid biosynthesis in Zephyranthes candida and therapeutic targets for LIHC.Planta. 2025 Jul 10;262(2):45. doi: 10.1007/s00425-025-04756-4. Planta. 2025. PMID: 40640563
-
Comprehensive Identification of HD-Zip Family Genes in Coix lacryma-jobi L. and Their Potential Roles in Response to Abiotic Stress.Plants (Basel). 2025 Apr 26;14(9):1318. doi: 10.3390/plants14091318. Plants (Basel). 2025. PMID: 40364347 Free PMC article.
References
-
- Latchman DS. Transcription factors: an overview. Int J Biochem Cell Biol. 1997;29:1305–12. - PubMed
-
- Amoutzias GD, Veron AS, Weiner IIIJ, Robinson-Rechavi M, Bornberg-Bauer E, Oliver SG, et al. One billion years of bZIP transcription factor evolution: conservation and change in dimerization and DNA-binding site specificity. Mol Biol Evol. 2007;24:827–35. - PubMed
-
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C-Z, Keddie J, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–10. - PubMed
-
- Klempnauer KH, Gonda TJ, Bishop JM. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell. 1982;31:453–63. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

