A multidimensional recommendation framework for identifying biological targets to aid the diagnosis and treatment of liver metastasis in patients with colorectal cancer
- PMID: 39449040
- PMCID: PMC11515508
- DOI: 10.1186/s12943-024-02155-z
A multidimensional recommendation framework for identifying biological targets to aid the diagnosis and treatment of liver metastasis in patients with colorectal cancer
Abstract
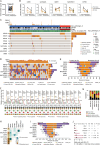
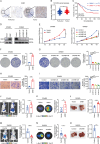
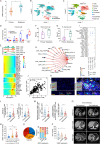
The quest to understand the molecular mechanisms of tumour metastasis and identify pivotal biomarkers for cancer therapy is increasing in importance. Single-omics analyses, constrained by their focus on a single biological layer, cannot fully elucidate the complexities of tumour molecular profiles and can thus overlook crucial molecular targets. In response to this limitation, we developed a multiobjective recommendation system (RJH-Metastasis 1.0) anchored in a multiomics knowledge graph to integrate genome, transcriptome, and proteome data and corroborative literature evidence and then conducted comprehensive analyses of colorectal cancer with liver metastasis (CRCLM). A total of 25 key genes significantly associated with CRCLM were recommended by our system, and GNB1, GATAD2A, GBP2, MACROD1, and EIF5B were further highlighted. Specifically, GNB1 presented fewer mutations but elevated RNA transcription and protein expression in CRCLM patients. The role of GNB1 in promoting the malignant behaviours of colon cancer cells was demonstrated via in vitro and in vivo studies. Aberrant expression of GNB1 could be regulated by METTL1-driven m7G modification. METTL1 knockdown decreased m7G modification in the 3' UTR of GNB1, increasing its mRNA transcription and translation during liver metastasis. Furthermore, GNB1 induced the formation of an immunosuppressive microenvironment by promoting the CLEC2C-KLRB1 interaction between memory B cells and KLRB1+PD-1+CD8+ cells. GNB1 expression and the efficacy of PD-1 antibody-based treatment in CRCLM patients were significantly correlated. In summary, our recommendation system can be used for effective exploration of key molecules in colorectal cancer, among which GNB1 was identified as a critical CRCLM promoter and immunotherapy biomarker in colorectal cancer patients.
Keywords: Colorectal cancer liver metastasis; GNB1; Immunotherapy; RNA modification; Recommendation system.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

