Identification of allele-specific KIV-2 repeats and impact on Lp(a) measurements for cardiovascular disease risk
- PMID: 39449055
- PMCID: PMC11515395
- DOI: 10.1186/s12920-024-02024-0
Identification of allele-specific KIV-2 repeats and impact on Lp(a) measurements for cardiovascular disease risk
Abstract
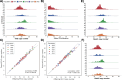
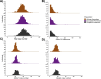
The abundance of Lp(a) protein holds significant implications for the risk of cardiovascular disease (CVD), which is directly impacted by the copy number (CN) of KIV-2, a 5.5 kbp sub-region. KIV-2 is highly polymorphic in the population and accurate analysis is challenging. In this study, we present the DRAGEN KIV-2 CN caller, which utilizes short reads. Data across 166 WGS show that the caller has high accuracy, compared to optical mapping and can further phase approximately 50% of the samples. We compared KIV-2 CN numbers to 24 previously postulated KIV-2 relevant SNVs, revealing that many are ineffective predictors of KIV-2 copy number. Population studies, including USA-based cohorts, showed distinct KIV-2 CN, distributions for European-, African-, and Hispanic-American populations and further underscored the limitations of SNV predictors. We demonstrate that the CN estimates correlate significantly with the available Lp(a) protein levels and that phasing is highly important.
Keywords: Cardiovascular disease; Copy number variation; Illumina; LPA; Next Generation Sequencing.
© 2024. The Author(s).
Conflict of interest statement
F.J.S receives research support from Illumina, PacBio and ONT. L.P. is funded from Genentech. E.N., M.R., E.R., J.H. and V.O. are employees from Illumina. J.R.B., X.C. and M.A.E. are employees from PacBio. V.K.M. is employed now at Genentech.
Figures




Update of
-
Identification of allele-specific KIV-2 repeats and impact on Lp(a) measurements for cardiovascular disease risk.bioRxiv [Preprint]. 2023 Apr 27:2023.04.24.538128. doi: 10.1101/2023.04.24.538128. bioRxiv. 2023. Update in: BMC Med Genomics. 2024 Oct 24;17(1):255. doi: 10.1186/s12920-024-02024-0. PMID: 37163057 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

