Age, sex, and cell type-resolved hypothalamic gene expression across the pubertal transition in mice
- PMID: 39449090
- PMCID: PMC11515584
- DOI: 10.1186/s13293-024-00661-9
Age, sex, and cell type-resolved hypothalamic gene expression across the pubertal transition in mice
Abstract
Background: The hypothalamus plays a central role in regulating puberty. However, our knowledge of the postnatal gene regulatory networks that control the pubertal transition in males and females is incomplete. Here, we investigate the age-, sex- and cell-type-specific gene regulation in the hypothalamus across the pubertal transition.
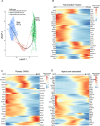
Methods: We used RNA-seq to profile hypothalamic gene expression in male and female mice at five time points spanning the onset of puberty (postnatal days (PD) 12, 22, 27, 32, and 37). By combining this data with hypothalamic single nuclei RNA-seq data from pre- and postpubertal mice, we assigned gene expression changes to their most likely cell types of origin. In our colony, pubertal onset occurs earlier in male mice, allowing us to focus on genes whose expression is dynamic across ages and offset between sexes, and to explore the bases of sex effects.
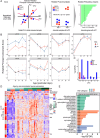
Results: Our age-by-sex pattern of expression enriched for biological pathways involved hormone production, neuronal activation, and glial maturation. Additionally, we inferred a robust expansion of oligodendrocytes precursor cells into mature oligodendrocytes spanning the prepubertal (PD12) to peri-pubertal (PD27) timepoints. Using spatial transcriptomic data from postpubertal mice, we observed the lateral hypothalamic area and zona incerta were the most oligodendrocyte-rich regions and that these cells expressed genes known to be involved in pubertal regulation.
Conclusion: Together, by incorporating multiple biological timepoints and using sex as a variable, we identified gene and cell-type changes that may participate in orchestrating the pubertal transition and provided a resource for future studies of postnatal hypothalamic gene regulation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

