Unraveling the glycosphingolipid metabolism by leveraging transcriptome-weighted network analysis on neuroblastic tumors
- PMID: 39449099
- PMCID: PMC11515559
- DOI: 10.1186/s40170-024-00358-y
Unraveling the glycosphingolipid metabolism by leveraging transcriptome-weighted network analysis on neuroblastic tumors
Abstract
Background: Glycosphingolipids (GSLs) are membrane lipids composed of a ceramide backbone linked to a glycan moiety. Ganglioside biosynthesis is a part of the GSL metabolism, which involves sequential reactions catalyzed by specific enzymes that in part have a poor substrate specificity. GSLs are deregulated in cancer, thus playing a role as potential biomarkers for personalized therapy or subtype classification. However, the analysis of GSL profiles is complex and requires dedicated technologies, that are currently not included in the commonly utilized high-throughput assays adopted in contexts such as molecular tumor boards.
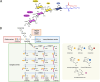
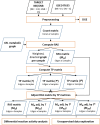
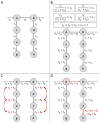
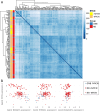
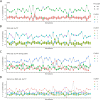
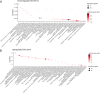
Methods: In this study, we developed a method to discriminate the enzyme activity among the four series of the ganglioside metabolism pathway by incorporating transcriptome data and topological information of the metabolic network. We introduced three adjustment options for reaction activity scores (RAS) and demonstrated their application in both exploratory and comparative analyses by applying the method on neuroblastic tumors (NTs), encompassing neuroblastoma (NB), ganglioneuroblastoma (GNB), and ganglioneuroma (GN). Furthermore, we interpreted the results in the context of earlier published GSL measurements in the same tumors.
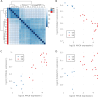
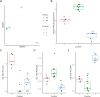
Results: By adjusting RAS values using a weighting scheme based on network topology and transition probabilities (TPs), the individual series of ganglioside metabolism can be differentiated, enabling a refined analysis of the GSL profile in NT entities. Notably, the adjustment method we propose reveals the differential engagement of the ganglioside series between NB and GNB. Moreover, MYCN gene expression, a well-known prognostic marker in NTs, appears to correlate with the expression of therapeutically relevant gangliosides, such as GD2. Using unsupervised learning, we identified subclusters within NB based on altered GSL metabolism.
Conclusion: Our study demonstrates the utility of adjusting RAS values in discriminating ganglioside metabolism subtypes, highlighting the potential for identifying novel cancer subgroups based on sphingolipid profiles. These findings contribute to a better understanding of ganglioside dysregulation in NT and may have implications for stratification and targeted therapeutic strategies in these tumors and other tumor entities with a deregulated GSL metabolism.
Keywords: GD2; Ganglioneuroblastoma; Ganglioneuroma; Ganglioside; Glycosphingolipids; Metabolic graph; Neuroblastoma; Reaction activity score.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Schnaar RL, Sandhoff R, Tiemeyer M, Kinoshita T. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al., editors. Glycosphingolipids. 4th ed. Cold Spring Harbor (NY); 2022. pp. 129–40. 10.1101/glycobiology.4e.11.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

