Dengue virus surveillance in Nepal yields the first on-site whole genome sequences of isolates from the 2022 outbreak
- PMID: 39449117
- PMCID: PMC11515306
- DOI: 10.1186/s12864-024-10879-x
Dengue virus surveillance in Nepal yields the first on-site whole genome sequences of isolates from the 2022 outbreak
Abstract
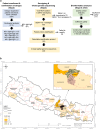
Background: The 4 serotypes of dengue virus (DENV1-4) can each cause potentially deadly dengue disease, and are spreading globally from tropical and subtropical areas to more temperate ones. Nepal provides a microcosm of this global phenomenon, having met each of these grim benchmarks. To better understand DENV transmission dynamics and spread into new areas, we chose to study dengue in Nepal and, in so doing, to build the onsite infrastructure needed to manage future, larger studies.
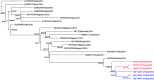
Methods and results: During the 2022 dengue season, we enrolled 384 patients presenting at a hospital in Kathmandu with dengue-like symptoms; 79% of the study participants had active or recent DENV infection (NS1 antigen and IgM). To identify circulating serotypes, we screened serum from 50 of the NS1+ participants by RT-PCR and identified DENV1, 2, and 3 - with DENV1 and 3 codominant. We also performed whole-genome sequencing of DENV, for the first time in Nepal, using our new on-site capacity. Sequencing analysis demonstrated the DENV1 and 3 genomes clustered with sequences reported from India in 2019, and the DENV2 genome clustered with a sequence reported from China in 2018.
Conclusion: These findings highlight DENV's geographic expansion from neighboring countries, identify China and India as the likely origin of the 2022 DENV cases in Nepal, and demonstrate the feasibility of building onsite capacity for more rapid genomic surveillance of circulating DENV. These ongoing efforts promise to protect populations in Nepal and beyond by informing the development and deployment of DENV drugs and vaccines in real time.
Keywords: Capacity building; Dengue outbreak; Dengue virus; Genomic surveillance; Whole genome sequencing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Update of
-
Dengue Virus Surveillance in Nepal Yields the First On-Site Whole Genome Sequences of Isolates from the 2022 Outbreak.bioRxiv [Preprint]. 2024 Jun 3:2024.06.02.597008. doi: 10.1101/2024.06.02.597008. bioRxiv. 2024. Update in: BMC Genomics. 2024 Oct 24;25(1):998. doi: 10.1186/s12864-024-10879-x. PMID: 38895410 Free PMC article. Updated. Preprint.
References
-
- O. WORLD WH, HEALTH ORGANIZATION. Dengue and severe dengue. https://www.who.int/news-room/questions-and-answers/item/dengue-and-seve... (accessed.
-
- Guzman MG, Harris E. Dengue, Lancet, vol. 385, no. 9966, pp. 453 – 65, Jan 31 2015, 10.1016/S0140-6736(14)60572-9 - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

