Subfamily C7 Raf-like kinases MRK1, RAF26, and RAF39 regulate immune homeostasis and stomatal opening in Arabidopsis thaliana
- PMID: 39449177
- PMCID: PMC11579443
- DOI: 10.1111/nph.20198
Subfamily C7 Raf-like kinases MRK1, RAF26, and RAF39 regulate immune homeostasis and stomatal opening in Arabidopsis thaliana
Abstract
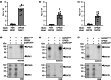
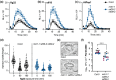
The calcium-dependent protein kinase CPK28 regulates several stress pathways in multiple plant species. Here, we aimed to discover CPK28-associated proteins in Arabidopsis thaliana. We used affinity-based proteomics and identified several potential CPK28 binding partners, including the C7 Raf-like kinases MRK1, RAF26, and RAF39. We used biochemistry, genetics, and physiological assays to gain insight into their function. We define redundant roles for these kinases in stomatal opening, immune-triggered reactive oxygen species (ROS) production, and resistance to a bacterial pathogen. We report that CPK28 associates with and trans-phosphorylates RAF26 and RAF39, and that MRK1, RAF26, and RAF39 are active kinases that localize to endomembranes. Although Raf-like kinases share some features with mitogen-activated protein kinase kinase kinases (MKKKs), we found that MRK1, RAF26, and RAF39 are unable to trans-phosphorylate any of the 10 Arabidopsis mitogen-activated protein kinase kinases (MKKs). Overall, our study suggests that C7 Raf-like kinases associate with and are phosphorylated by CPK28, function redundantly in stomatal opening and immunity, and possess substrate specificities distinct from canonical MKKKs.
Keywords: Arabidopsis thaliana; C7 Raf‐like kinase; CPK28; MRK1; RAF26; RAF39; immunity; stomata.
© 2024 The Author(s). New Phytologist © 2024 New Phytologist Foundation.
Conflict of interest statement
None declared.
Figures



References
-
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nature Biotechnology 37: 420–423. - PubMed
-
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu W‐L, Gomez‐Gomez L, Boller T, Ausubel FM, Sheen J. 2002. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

