Epigenome Mapping in Quiescent Cells Reveals a Key Role for H3K4me3 in Regulation of RNA Polymerase II Activity
- PMID: 39449363
- PMCID: PMC11503321
- DOI: 10.3390/epigenomes8040039
Epigenome Mapping in Quiescent Cells Reveals a Key Role for H3K4me3 in Regulation of RNA Polymerase II Activity
Abstract
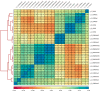
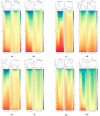
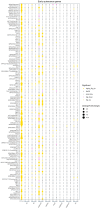
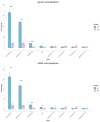
(1) Background: Quiescent cells are those that have stopped dividing and show strongly reduced levels of gene expression during dormancy. In response to appropriate signals, the cells can wake up and start growing again. Many histone modifications are regulated in quiescence, but their exact functions remain to be determined. (2) Methods: Here, we map the different histone modifications, H3K4me3, H3K9ac, H3K9me2, and H3K9me3, and the histone variant H2A.Z, comparing vegetative and quiescent fission yeast (S. pombe) cells. We also map histone H3 as a control and RNA polymerase II (phosphorylated at S2 and S5) to enable comparisons of their occupancies within genes. We use ChIP-seq methodology and several different bioinformatics tools. (3) Results: The histone modification mapping data show that H3K4me3 changes stand out as being the most significant. Changes in occupancy of histone variant H2A.Z were also significant, consistent with earlier studies. Regarding gene expression changes in quiescence, we found that changes in mRNA levels were associated with changes in occupancy of RNA polymerase II (S2 and S5). Analysis of quiescence genes showed that increased H3K4me3 levels and RNA polymerase II occupancy were super-significant in a small set of core quiescence genes that are continuously upregulated during dormancy. We demonstrate that several of these genes were require Set1C/COMPASS activity for their strong induction during quiescence. (4) Conclusions: Our results imply that regulation of gene expression in quiescent cells involves epigenome changes with a key role for H3K4me3 in regulation of RNA polymerase II activity, and that different gene activation mechanisms control early and core quiescence genes. Thus, our data give further insights into important epigenome changes in quiescence using fission yeast as an experimental model.
Keywords: G0 arrest; H3K4me3; RNA polymerase II; Set1C/COMPASS; cellular quiescence; fission yeast; histone modifications; regulation of gene expression.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures








Similar articles
-
LEO1 Is Required for Efficient Entry into Quiescence, Control of H3K9 Methylation and Gene Expression in Human Fibroblasts.Biomolecules. 2023 Nov 17;13(11):1662. doi: 10.3390/biom13111662. Biomolecules. 2023. PMID: 38002344 Free PMC article.
-
An essential role for the Ino80 chromatin remodeling complex in regulation of gene expression during cellular quiescence.Chromosome Res. 2023 Apr 12;31(2):14. doi: 10.1007/s10577-023-09723-x. Chromosome Res. 2023. PMID: 37043046 Free PMC article.
-
Distinct histone methylation and transcription profiles are established during the development of cellular quiescence in yeast.BMC Genomics. 2017 Jan 26;18(1):107. doi: 10.1186/s12864-017-3509-9. BMC Genomics. 2017. PMID: 28122508 Free PMC article.
-
Transcriptional reprogramming in cellular quiescence.RNA Biol. 2017 Jul 3;14(7):843-853. doi: 10.1080/15476286.2017.1327510. Epub 2017 May 12. RNA Biol. 2017. PMID: 28497998 Free PMC article. Review.
-
Is There a Histone Code for Cellular Quiescence?Front Cell Dev Biol. 2021 Oct 29;9:739780. doi: 10.3389/fcell.2021.739780. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34778253 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

