Remodeling tumor microenvironment using pH-sensitive biomimetic co-delivery of TRAIL/R848 liposomes against colorectal cancer
- PMID: 39449815
- PMCID: PMC11497182
- DOI: 10.32604/or.2024.045564
Remodeling tumor microenvironment using pH-sensitive biomimetic co-delivery of TRAIL/R848 liposomes against colorectal cancer
Abstract
Background: Despite significant advancements in the development of anticancer therapies over the past few decades, the clinical management of colorectal cancer remains a challenging task. This study aims to investigate the inhibitory effects of cancer-targeting liposomes against colorectal cancer.
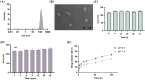
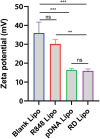
Materials and methods: Liposomes consisting of 3β-[N-(N', N'-dimethylamino ethane)carbamoyl]-cholesterol (DC-CHOL), cholesterol (CHOL), and dioleoylphosphatidylethanolamine (DOPE) at a molar ratio of 1:1:0.5 were created and used as carriers to deliver an apoptosis-inducing plasmid encoding the tumor necrosis factor-related apoptosis-inducing ligand (pTRAIL) gene, along with the toll-like receptor (TLR7) agonist Rsiquimod (R848). The rationale behind this design is that pTRAIL can trigger cancer cell apoptosis by activating the DR4/5 receptor, while R848 can stimulate the immune microenvironment.
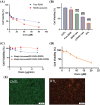
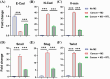
Results: Experimental results demonstrated the synergistic effects of R848 and pTRAIL encapsulated by liposomes (RTL) in suppressing the proliferation of colorectal cancer cells. Moreover, further in vivo investigations revealed the strong anti-tumor efficacy of RTL in xenograft and orthotropic in situ models of colorectal cancer.
Conclusions: These findings collectively highlight the therapeutic potential of R848/pTRAIL-loaded liposomes in the treatment of colorectal cancer.
Keywords: Colorectal cancer; Plasmid TRAIL (pTRAIL); R848; Tumor-associated macrophages.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Combination of TRAIL and actinomycin D liposomes enhances antitumor effect in non-small cell lung cancer.Int J Nanomedicine. 2012;7:1449-60. doi: 10.2147/IJN.S24711. Epub 2012 Mar 19. Int J Nanomedicine. 2012. PMID: 22619505 Free PMC article.
-
Prediction of proapoptotic anticancer therapeutic response in vivo based on cell death visualization and TRAIL death ligand-receptor interaction.Cancer Biol Ther. 2011 Aug 15;12(4):335-48. doi: 10.4161/cbt.12.4.17174. Epub 2011 Aug 15. Cancer Biol Ther. 2011. PMID: 21785270
-
Breathing New Life into TRAIL for Breast Cancer Therapy: Co-Delivery of pTRAIL and Complementary siRNAs Using Lipopolymers.Hum Gene Ther. 2019 Dec;30(12):1531-1546. doi: 10.1089/hum.2019.096. Epub 2019 Oct 29. Hum Gene Ther. 2019. PMID: 31547718
-
Complexes containing cationic and anionic pH-sensitive liposomes: comparative study of factors influencing plasmid DNA gene delivery to tumors.Int J Nanomedicine. 2013;8:1573-93. doi: 10.2147/IJN.S42800. Epub 2013 Apr 22. Int J Nanomedicine. 2013. PMID: 23637529 Free PMC article.
-
Lessons from TRAIL-resistance mechanisms in colorectal cancer cells: paving the road to patient-tailored therapy.Drug Resist Updat. 2004 Dec;7(6):345-58. doi: 10.1016/j.drup.2004.11.002. Epub 2005 Jan 8. Drug Resist Updat. 2004. PMID: 15790545 Review.
Cited by
-
Innovations in Cancer Therapy: Endogenous Stimuli-Responsive Liposomes as Advanced Nanocarriers.Pharmaceutics. 2025 Feb 13;17(2):245. doi: 10.3390/pharmaceutics17020245. Pharmaceutics. 2025. PMID: 40006612 Free PMC article. Review.
References
-
- Vernon, S. W., McQueen, A., Holland, J., Breitbart, W., Jacobsen, P.et al. (2010). Colorectal cancer screening. Hematology-Oncology Clinics of North America , 3, 393–414.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
