Phenotyping of Panicle Number and Shape in Rice Breeding Materials Based on Unmanned Aerial Vehicle Imagery
- PMID: 39449974
- PMCID: PMC11499587
- DOI: 10.34133/plantphenomics.0265
Phenotyping of Panicle Number and Shape in Rice Breeding Materials Based on Unmanned Aerial Vehicle Imagery
Abstract
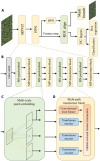
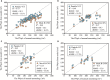
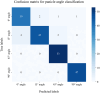
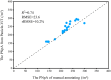
The number of panicles per unit area (PNpA) is one of the key factors contributing to the grain yield of rice crops. Accurate PNpA quantification is vital for breeding high-yield rice cultivars. Previous studies were based on proximal sensing with fixed observation platforms or unmanned aerial vehicles (UAVs). The near-canopy images produced in these studies suffer from inefficiency and complex image processing pipelines that require manual image cropping and annotation. This study aims to develop an automated, high-throughput UAV imagery-based approach for field plot segmentation and panicle number quantification, along with a novel classification method for different panicle types, enhancing PNpA quantification at the plot level. RGB images of the rice canopy were efficiently captured at an altitude of 15 m, followed by image stitching and plot boundary recognition via a mask region-based convolutional neural network (Mask R-CNN). The images were then segmented into plot-scale subgraphs, which were categorized into 3 growth stages. The panicle vision transformer (Panicle-ViT), which integrates a multipath vision transformer and replaces the Mask R-CNN backbone, accurately detects panicles. Additionally, the Res2Net50 architecture classified panicle types with 4 angles of 0°, 15°, 45°, and 90°. The results confirm that the performance of Plot-Seg is comparable to that of manual segmentation. Panicle-ViT outperforms the traditional Mask R-CNN across all the datasets, with the average precision at 50% intersection over union (AP50) improved by 3.5% to 20.5%. The PNpA quantification for the full dataset achieved superior performance, with a coefficient of determination (R 2) of 0.73 and a root mean square error (RMSE) of 28.3, and the overall panicle classification accuracy reached 94.8%. The proposed approach enhances operational efficiency and automates the process from plot cropping to PNpA prediction, which is promising for accelerating the selection of desired traits in rice breeding.
Copyright © 2024 Xuqi Lu et al.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures







References
-
- Seck PA, Diagne A, Mohanty S, Wopereis MCS. Crops that feed the world 7: Rice. Food Secur. 2012;4(1):7–24.
-
- Kolukisaoglu U, Thurow K. Future and frontiers of automated screening in plant sciences. Plant Sci. 2010;178(6):476–484.
-
- Yang W, Duan L, Chen G, Xiong L, Liu Q. Plant phenomics and high-throughput phenotyping: Accelerating rice functional genomics using multidisciplinary technologies. Curr Opin Plant Biol. 2013;16(2):180–187. - PubMed
-
- Kim HY, Ko J, Kang S, Tenhunen J. Impacts of climate change on paddy rice yield in a temperate climate. Glob Chang Biol. 2013;19(2):548–562. - PubMed
LinkOut - more resources
Full Text Sources

