The genome sequence of the Currant Clearwing moth, Synanthedon tipuliformis (Clerck, 1759)
- PMID: 39449983
- PMCID: PMC11499843
- DOI: 10.12688/wellcomeopenres.19647.2
The genome sequence of the Currant Clearwing moth, Synanthedon tipuliformis (Clerck, 1759)
Abstract
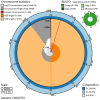
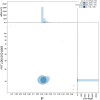
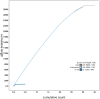
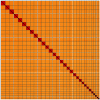
We present a genome assembly from an individual male Synanthedon tipuliformis (the Currant Clearwing; Arthropoda; Insecta; Lepidoptera; Sesiidae). The genome sequence is 295.8 megabases in span. Most of the assembly (99.98%) is scaffolded into 31 chromosomal pseudomolecules, including the Z sex chromosome. The mitochondrial genome has also been assembled and is 27.05 kilobases in length. Gene annotation of this assembly on Ensembl identified 11,878 protein-coding genes.
Keywords: Currant Clearwing; Lepidoptera; Synanthedon tipuliformis; chromosomal; genome sequence.
Copyright: © 2024 Boyes D et al.
Conflict of interest statement
No competing interests were disclosed.
Figures

References
LinkOut - more resources
Full Text Sources

