Pre-vaccination transcriptomic profiles of immune responders to the MUC1 peptide vaccine for colon cancer prevention
- PMID: 39450169
- PMCID: PMC11499122
- DOI: 10.3389/fimmu.2024.1437391
Pre-vaccination transcriptomic profiles of immune responders to the MUC1 peptide vaccine for colon cancer prevention
Abstract
Introduction: Self-antigens abnormally expressed on tumors, such as MUC1, have been targeted by therapeutic cancer vaccines. We recently assessed in two clinical trials in a preventative setting whether immunity induced with a MUC1 peptide vaccine could reduce high colon cancer risk in individuals with a history of premalignant colon adenomas. In both trials, there were immune responders and non-responders to the vaccine.
Methods: Here we used PBMC pre-vaccination and 2 weeks after the first vaccine of responders and non-responders selected from both trials to identify early biomarkers of immune response involved in long-term memory generation and prevention of adenoma recurrence. We performed flow cytometry, phosflow, and differential gene expression analyses on PBMCs collected from MUC1 vaccine responders and non-responders pre-vaccination and two weeks after the first of three vaccine doses.
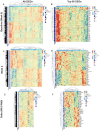
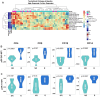
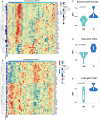
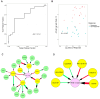
Results: MUC1 vaccine responders had higher frequencies of CD4 cells pre-vaccination, increased expression of CD40L on CD8 and CD4 T-cells, and a greater increase in ICOS expression on CD8 T-cells. Differential gene expression analysis revealed that iCOSL, PI3K AKT MTOR, and B-cell signaling pathways are activated early in response to the MUC1 vaccine. We identified six specific transcripts involved in elevated antigen presentation, B-cell activation, and NF-κB1 activation that were directly linked to finding antibody response at week 12. Finally, a model using these transcripts was able to predict non-responders with accuracy.
Discussion: These findings suggest that individuals who can be predicted to respond to the MUC1 vaccine, and potentially other vaccines, have greater readiness in all immune compartments to present and respond to antigens. Predictive biomarkers of MUC1 vaccine response may lead to more effective vaccines tailored to individuals with high risk for cancer but with varying immune fitness.
Keywords: MUC1; cancer vaccine; colon cancer; colorectal adenoma; serological response; transcriptomics.
Copyright © 2024 Cameron, Raghu, Richardson, Zagore, Tamilselvan, Golden, Cartwright, Schoen, Finn, Benos and Cameron.
Conflict of interest statement
Author RS reports support from Freenome, Exact Sciences, and Immunovia during the conduct of the study. Author OF reports personal fees from PDS Biotech, Invectys, Immodulon, and Ardigen outside the submitted work. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


Update of
-
Pre-vaccination transcriptomic profiles of immune responders to the MUC1 peptide vaccine for colon cancer prevention.medRxiv [Preprint]. 2024 May 10:2024.05.09.24305336. doi: 10.1101/2024.05.09.24305336. medRxiv. 2024. Update in: Front Immunol. 2024 Oct 10;15:1437391. doi: 10.3389/fimmu.2024.1437391. PMID: 38766010 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

