SHARPIN is a novel gene of colorectal cancer that promotes tumor growth potentially via inhibition of p53 expression
- PMID: 39450547
- PMCID: PMC11542962
- DOI: 10.3892/ijo.2024.5701
SHARPIN is a novel gene of colorectal cancer that promotes tumor growth potentially via inhibition of p53 expression
Abstract
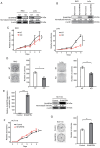
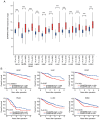
Colorectal cancer (CRC) is widely prevalent and represents a significant contributor to global cancer‑related mortality. There remains a pressing demand for advancements in CRC treatment modalities. The E3 ubiquitin ligase is a critical enzyme involved in modulating protein expression levels via posttranslational ubiquitin‑mediated proteolysis, and it is reportedly involved in the progression of various cancers, making it a target of recent interest in anticancer therapy. In the present study, using comprehensive expression analysis involving spatial transcriptomic analysis with single‑cell RNA sequencing in clinical CRC datasets, the ubiquitin‑associated protein Shank‑associated RH domain interactor (SHARPIN) was identified, located on amplified chromosome 8q, which could promote CRC progression. SHARPIN was found to be upregulated in tumor cells, with elevated expression observed in tumor tissues. This heightened expression of SHARPIN was positively associated with lymphatic invasion and served as an independent predictor of a poor prognosis in patients with CRC. In vitro and in vivo analyses using SHARPIN‑overexpressing or ‑knockout CRC cells revealed that SHARPIN overexpression upregulated MDM2, resulting in the downregulation of p53, while SHARPIN silencing or knockout downregulated MDM2, leading to p53 upregulation, which affects cell cycle progression, tumor cell apoptosis and tumor growth in CRC. Furthermore, SHARPIN was found to be overexpressed in several cancer types, exerting significant effects on survival outcomes. In conclusion, SHARPIN represents a newly identified novel gene with the potential to promote tumor growth following apoptosis inhibition and cell cycle progression in part by inhibiting p53 expression via MDM2 upregulation; therefore, SHARPIN represents a potential therapeutic target for CRC.
Keywords: colorectal cancer; novel gene; p53; shank‑associated RH domain interactor; ubiquitin‑proteasome system.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

