Single-cell Sequencing of Circulating Human Plasmablasts during Staphylococcus aureus Bacteremia
- PMID: 39451041
- PMCID: PMC7616744
- DOI: 10.4049/jimmunol.2300858
Single-cell Sequencing of Circulating Human Plasmablasts during Staphylococcus aureus Bacteremia
Abstract
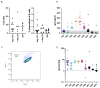
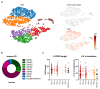
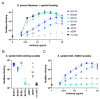
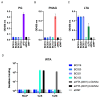
Staphylococcus aureus is the major cause of healthcare-associated infections, including life-threatening conditions as bacteremia, endocarditis, and implant-associated infections. Despite adequate antibiotic treatment, the mortality of S. aureus bacteremia remains high. This calls for different strategies to treat this infection. In past years, sequencing of Ab repertoires from individuals previously exposed to a pathogen emerged as a successful method to discover novel therapeutic monoclonal Abs and understand circulating B cell diversity during infection. In this paper, we collected peripheral blood from 17 S. aureus bacteremia patients to study circulating plasmablast responses. Using single-cell transcriptome gene expression combined with sequencing of variable heavy and light Ig genes, we retrieved sequences from >400 plasmablasts revealing a high diversity with >300 unique variable heavy and light sequences. More than 200 variable sequences were synthesized to produce recombinant IgGs that were analyzed for binding to S. aureus whole bacterial cells. This revealed four novel monoclonal Abs that could specifically bind to the surface of S. aureus in the absence of Ig-binding surface SpA. Interestingly, three of four mAbs showed cross-reactivity with Staphylococcus epidermidis. Target identification revealed that the S. aureus-specific mAb BC153 targets wall teichoic acid, whereas cross-reactive mAbs BC019, BC020, and BC021 target lipoteichoic acid. All mAbs could induce Fc-dependent phagocytosis of staphylococci by human neutrophils. Altogether, we characterize the active B cell responses to S. aureus in infected patients and identify four functional mAbs against the S. aureus surface, of which three cross-react with S. epidermidis.
Copyright © 2024 by The American Association of Immunologists, Inc.
Figures

References
-
- Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–32. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

