Identification of the Optimal Quantitative RT-PCR Reference Gene for Paper Mulberry (Broussonetia papyrifera)
- PMID: 39451520
- PMCID: PMC11506246
- DOI: 10.3390/cimb46100640
Identification of the Optimal Quantitative RT-PCR Reference Gene for Paper Mulberry (Broussonetia papyrifera)
Abstract
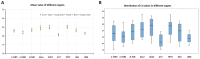
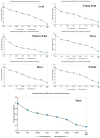
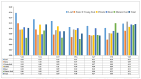
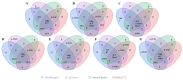
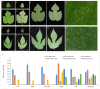
Paper Mulberry (Broussonetia papyrifera) possesses medicinal, economic, and ecological significance and is extensively used for feed production, papermaking, and ecological restoration due to its ease of propagation, rapid growth rate, and strong stress resistance. The recent completion of the sequencing of the Paper Mulberry genome has prompted further research into the genetic breeding and molecular biology of this important species. A highly stable reference gene is essential to enhance the quantitative analysis of functional genes in Paper Mulberry; however, none has been identified. Accordingly, in this study, the leaves, stems, roots, petioles, young fruits, and mature fruits of Paper Mulberry plants were selected as experimental materials, and nine candidate reference genes, namely, α-TUB1, α-TUB2, β-TUB, H2A, ACT, DnaJ, UBQ, CDC2, and TIP41, were identified by RT-qPCR. Their stability was assessed using the geNorm, Normfinder, Delta Ct, BestKeeper, and RefFinder algorithms, identifying ACT and UBQ as showing the greatest stability. The expression of BpMYB090, which regulates the production of trichomes, was examined in the leaves of plants of the wild type (which have more trichomes) and mutant (which have fewer trichomes) at various developmental stages to validate the results of this study. As a result, their identification addresses a critical gap in the field of Paper Mulberry research, providing a solid foundation for future research that will concentrate on the characterization of pertinent functional genes in this economically valuable species.
Keywords: Broussonetia papyrifera; real-time quantitative PCR; reference gene.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Screening and Validation of Appropriate Reference Genes for Real-Time Quantitative PCR under PEG, NaCl and ZnSO4 Treatments in Broussonetia papyrifera.Int J Mol Sci. 2023 Oct 11;24(20):15087. doi: 10.3390/ijms242015087. Int J Mol Sci. 2023. PMID: 37894768 Free PMC article.
-
A Chromosome-Scale Genome Assembly of Paper Mulberry (Broussonetia papyrifera) Provides New Insights into Its Forage and Papermaking Usage.Mol Plant. 2019 May 6;12(5):661-677. doi: 10.1016/j.molp.2019.01.021. Epub 2019 Feb 26. Mol Plant. 2019. PMID: 30822525
-
A Selection of Reliable Reference Genes for Gene Expression Analysis in the Female and Male Flowers of Salix suchowensis.Plants (Basel). 2022 Feb 27;11(5):647. doi: 10.3390/plants11050647. Plants (Basel). 2022. PMID: 35270117 Free PMC article.
-
Bioinformatics analysis and function prediction of NBS-LRR gene family in Broussonetia papyrifera.Biotechnol Lett. 2023 Jan;45(1):13-31. doi: 10.1007/s10529-022-03318-y. Epub 2022 Nov 10. Biotechnol Lett. 2023. PMID: 36357714 Review.
-
Medicinal Potential of Broussonetia papyrifera: Chemical Composition and Biological Activity Analysis.Plants (Basel). 2025 Feb 8;14(4):523. doi: 10.3390/plants14040523. Plants (Basel). 2025. PMID: 40006781 Free PMC article. Review.
Cited by
-
The Mulberry WRKY Transcription Factor MaWRKYIIc7 Participates in Regulating Plant Drought Stress Tolerance.Int J Mol Sci. 2025 Feb 17;26(4):1714. doi: 10.3390/ijms26041714. Int J Mol Sci. 2025. PMID: 40004176 Free PMC article.
-
Transcriptomic Analysis of Broussonetia papyrifera Fruit Under Manganese Stress and Mining of Flavonoid Synthesis Genes.Plants (Basel). 2025 Mar 12;14(6):883. doi: 10.3390/plants14060883. Plants (Basel). 2025. PMID: 40265799 Free PMC article.
References
-
- Morgan E.C., Overholt W.A. Wildland Weeds: Paper Mulberry, Broussonetia papyrifera. University of Florida, IFAS; Gainesville, FL, USA: 2004. Extension: 2.
-
- Li G.Y., Hu N., Ding D.X., Zheng J.F., Liu Y.L., Wang Y.D., Nie X.Q. Screening of plant species for phytoremediation of uranium, thorium, barium, nickel, strontium and lead contaminated soils from a uranium mill tailings repository in South China. Bull. Environ. Contam. Toxicol. 2011;86:646–652. doi: 10.1007/s00128-011-0291-2. - DOI - PubMed
-
- Wang J., Liu J., Peng X., Ni Z., Wang G., Shen S. Applied hybrid Paper Mulberry in ecological virescence of the coastal saline. Tianjin Agr. Sci. 2014;20:95–101.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

