Early Cervical Cancer Diagnosis with SWIN-Transformer and Convolutional Neural Networks
- PMID: 39451609
- PMCID: PMC11506970
- DOI: 10.3390/diagnostics14202286
Early Cervical Cancer Diagnosis with SWIN-Transformer and Convolutional Neural Networks
Abstract
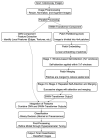
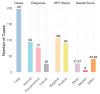
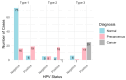
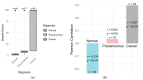
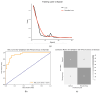
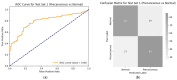
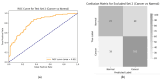
Introduction: Early diagnosis of cervical cancer at the precancerous stage is critical for effective treatment and improved patient outcomes. Objective: This study aims to explore the use of SWIN Transformer and Convolutional Neural Network (CNN) hybrid models combined with transfer learning to classify precancerous colposcopy images. Methods: Out of 913 images from 200 cases obtained from the Colposcopy Image Bank of the International Agency for Research on Cancer, 898 met quality standards and were classified as normal, precancerous, or cancerous based on colposcopy and histopathological findings. The cases corresponding to the 360 precancerous images, along with an equal number of normal cases, were divided into a 70/30 train-test split. The SWIN Transformer and CNN hybrid model combines the advantages of local feature extraction by CNNs with the global context modeling by SWIN Transformers, resulting in superior classification performance and a more automated process. The hybrid model approach involves enhancing image quality through preprocessing, extracting local features with CNNs, capturing the global context with the SWIN Transformer, integrating these features for classification, and refining the training process by tuning hyperparameters. Results: The trained model achieved the following classification performances on fivefold cross-validation data: a 94% Area Under the Curve (AUC), an 88% F1 score, and 87% accuracy. On two completely independent test sets, which were never seen by the model during training, the model achieved an 80% AUC, a 75% F1 score, and 75% accuracy on the first test set (precancerous vs. normal) and an 82% AUC, a 78% F1 score, and 75% accuracy on the second test set (cancer vs. normal). Conclusions: These high-performance metrics demonstrate the models' effectiveness in distinguishing precancerous from normal colposcopy images, even with modest datasets, limited data augmentation, and the smaller effect size of precancerous images compared to malignant lesions. The findings suggest that these techniques can significantly aid in the early detection of cervical cancer at the precancerous stage.
Keywords: SWIN Transformer; cancer screening; cervical cancer; colposcopy images; convolutional neural networks (CNN); early diagnosis; histopathology; hybrid models; medical image classification; precancerous lesions; transfer learning.
Conflict of interest statement
Author Seid Muhie was employed by Enkoy LLC, the remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Enhanced Pneumonia Detection in Chest X-Rays Using Hybrid Convolutional and Vision Transformer Networks.Curr Med Imaging. 2025;21:e15734056326685. doi: 10.2174/0115734056326685250101113959. Curr Med Imaging. 2025. PMID: 39806960
-
Swin-GA-RF: genetic algorithm-based Swin Transformer and random forest for enhancing cervical cancer classification.Front Oncol. 2024 Jul 19;14:1392301. doi: 10.3389/fonc.2024.1392301. eCollection 2024. Front Oncol. 2024. PMID: 39099689 Free PMC article.
-
Classification of Mobile-Based Oral Cancer Images Using the Vision Transformer and the Swin Transformer.Cancers (Basel). 2024 Feb 29;16(5):987. doi: 10.3390/cancers16050987. Cancers (Basel). 2024. PMID: 38473348 Free PMC article.
-
Efficient brain tumor segmentation using Swin transformer and enhanced local self-attention.Int J Comput Assist Radiol Surg. 2024 Feb;19(2):273-281. doi: 10.1007/s11548-023-03024-8. Epub 2023 Oct 5. Int J Comput Assist Radiol Surg. 2024. PMID: 37796413 Review.
-
An improved feature extraction algorithm for robust Swin Transformer model in high-dimensional medical image analysis.Comput Biol Med. 2025 Apr;188:109822. doi: 10.1016/j.compbiomed.2025.109822. Epub 2025 Feb 20. Comput Biol Med. 2025. PMID: 39983364 Review.
Cited by
-
Leveraging swin transformer with ensemble of deep learning model for cervical cancer screening using colposcopy images.Sci Rep. 2025 Mar 6;15(1):7900. doi: 10.1038/s41598-025-90415-3. Sci Rep. 2025. PMID: 40050635 Free PMC article.
References
-
- WHO World Health Organization Cervical Cancer: Key Facts. [(accessed on 29 August 2024)]. Available online: https://www.who.int/news-room/fact-sheets/detail/cervical-cancer.
-
- International Agency for Research on Cancer (IARC) New Estimates of the Number of Cervical Cancer Cases and Deaths That Could Be Averted in Low-Income and Lower-Middle-Income Countries by Scale-up of Screening and Vaccination Activities. 2024. [(accessed on 24 April 2024)]. Available online: https://www.iarc.who.int/news-events/new_estimates_cervical_cancer_cases...
-
- Ferlay J., Ervik M., Lam F., Colombet M., Mery L., Piñeros M., Znaor A., Soerjomataram I., Bray F. Global Cancer Observatory: Cancer Today 2019. [(accessed on 10 August 2024)]. Available online: https://publications.iarc.fr/Databases/Iarc-Cancerbases/Cancer-Today-Pow....
LinkOut - more resources
Full Text Sources
Miscellaneous

