Machine Learning Diagnostic Model for Early Stage NSTEMI: Using hs-cTnI 1/2h Changes and Multiple Cardiovascular Biomarkers
- PMID: 39451645
- PMCID: PMC11506866
- DOI: 10.3390/diagnostics14202322
Machine Learning Diagnostic Model for Early Stage NSTEMI: Using hs-cTnI 1/2h Changes and Multiple Cardiovascular Biomarkers
Abstract
Background: This study demonstrates differences in the distribution of multiple cardiovascular biomarkers between non-ST-segment elevation myocardial infarction (NSTEMI) and unstable angina (UA) patients. Diagnostic machine learning predictive models measured at the time of admission and 1/2 h post-admission, achieving competitive diagnostic predictive results.
Objective: This study aims to explore the diagnostic value of changes in high-sensitivity cardiac troponin I (hs-cTnI) levels in patients with suspected NSTEMI.
Methods: A total of 267 patients presented with chest pain, requiring confirmation of acute coronary syndrome (ACS) subtypes (NSTEMI vs. UA). Hs-cTnI and other cardiac markers, such as creatine kinase-MB (CK-MB) and Myoglobin (Myo), were analyzed. Machine learning techniques were employed to assess the application of hs-cTnI level changes in the clinical diagnosis of NSTEMI.
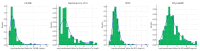
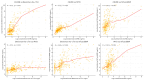
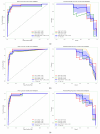
Results: Levels of CK-MB, Myo, hs-cTnI measured at admission, hs-cTnI measured 1-2 h after admission, and NT-proBNP in NSTEMI patients were significantly higher than those in UA patients (p < 0.001). There was a positive correlation between hs-cTnI and CK-MB, as well as Myo (R = 0.72, R = 0.51, R = 0.60). The optimal diagnostic model, Hybiome_1/2h, demonstrated an F1-Score of 0.74, an AUROC of 0.96, and an AP of 0.89.
Conclusions: This study confirms the significant value of hs-cTnI as a sensitive marker of myocardial injury in the diagnosis of NSTEMI. Continuous monitoring of hs-cTnI levels enhances the accuracy of distinguishing NSTEMI from UA. The models indicate that the Hybiome hs-cTnI assays perform comparably well to the Beckman assays in predicting NSTEMI. Moreover, incorporating hs-cTnI measurements taken 1-2 h post-admission significantly enhances the model's effectiveness.
Keywords: NSTEMI; diagnostic model; machine learning.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- Chiang C.H., Chiang C.H., Pickering J.W., Stoyanov K.M., Chew D.P., Neumann J.T., Ojeda F., Sörensen N.A., Su K.Y., Kavsak P., et al. Performance of the European Society of Cardiology 0/1-Hour, 0/2-Hour, and 0/3-Hour Algorithms for Rapid Triage of Acute Myocardial Infarction: An International Collaborative Meta-Analysis. Ann. Intern. Med. 2022;175:101–113. doi: 10.7326/M21-1499. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

