Aptamer-Mediated Electrochemical Detection of SARS-CoV-2 Nucleocapsid Protein in Saliva
- PMID: 39451684
- PMCID: PMC11505747
- DOI: 10.3390/bios14100471
Aptamer-Mediated Electrochemical Detection of SARS-CoV-2 Nucleocapsid Protein in Saliva
Abstract
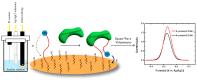
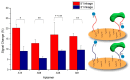
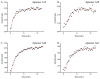
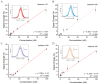
Gold standard detection of SARS-CoV-2 by reverse transcription quantitative PCR (RT-qPCR) can achieve ultrasensitive viral detection down to a few RNA copies per sample. Yet, the lengthy detection and labor-intensive protocol limit its effectiveness in community screening. In view of this, a structural switching electrochemical aptamer-based biosensor (E-AB) targeting the SARS-CoV-2 nucleocapsid (N) protein was developed. Four N protein-targeting aptamers were characterized on an electrochemical cell configuration using square wave voltammetry (SWV). The sensor was investigated in an artificial saliva matrix optimizing the aptamer anchoring orientation, SWV interrogation frequency, and target incubation time. Rapid detection of the N protein was achieved within 5 min at a low nanomolar limit of detection (LOD) with high specificity. Specific N protein detection was also achieved in simulated positive saliva samples, demonstrating its feasibility for saliva-based rapid diagnosis. Further research will incorporate novel signal amplification strategies to improve sensitivity for early diagnosis.
Keywords: SARS-CoV-2 nucleocapsid protein; electrochemical sensor; structural switching aptamer.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
Voltammetric-based immunosensor for the detection of SARS-CoV-2 nucleocapsid antigen.Mikrochim Acta. 2021 May 26;188(6):199. doi: 10.1007/s00604-021-04867-1. Mikrochim Acta. 2021. PMID: 34041585 Free PMC article.
-
Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: A reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva.Biosens Bioelectron. 2021 Jan 1;171:112686. doi: 10.1016/j.bios.2020.112686. Epub 2020 Oct 3. Biosens Bioelectron. 2021. PMID: 33086175 Free PMC article.
-
Optimized aptamer-based next generation biosensor for the ultra-sensitive determination of SARS-CoV-2 S1 protein in saliva samples.Int J Biol Macromol. 2024 Nov;281(Pt 1):136233. doi: 10.1016/j.ijbiomac.2024.136233. Epub 2024 Oct 1. Int J Biol Macromol. 2024. PMID: 39362419
-
Dual structure-switching aptamer-mediated signal amplification cascade for SARS-CoV-2 detection.Biosens Bioelectron. 2024 Sep 1;259:116375. doi: 10.1016/j.bios.2024.116375. Epub 2024 May 9. Biosens Bioelectron. 2024. PMID: 38749283
-
A universal three-dimensional hydrogel electrode for electrochemical detection of SARS-CoV-2 nucleocapsid protein and hydrogen peroxide.Biosens Bioelectron. 2024 Sep 1;259:116355. doi: 10.1016/j.bios.2024.116355. Epub 2024 May 1. Biosens Bioelectron. 2024. PMID: 38754196
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

