Proteomic Analysis of Human Serum Proteins Adsorbed onto Collagen Barrier Membranes
- PMID: 39452600
- PMCID: PMC11508515
- DOI: 10.3390/jfb15100302
Proteomic Analysis of Human Serum Proteins Adsorbed onto Collagen Barrier Membranes
Abstract
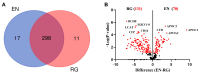
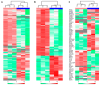
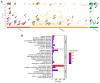
Collagen barrier membranes are frequently used in guided tissue and bone regeneration. The aim of this study was to analyze the signature of human serum proteins adsorbed onto collagen membranes using a novel protein extraction method combined with mass spectrometry. Native porcine-derived collagen membranes (Geistlich Bio-Gide®, Wolhusen, Switzerland) were exposed to pooled human serum in vitro and, after thorough washing, subjected to protein extraction either in conjunction with protein enrichment or via a conventional surfactant-based method. The extracted proteins were analyzed via liquid chromatography with tandem mass spectrometry. Bioinformatic analysis of global profiling, gene ontology, and functional enrichment of the identified proteins was performed. Overall, a total of 326 adsorbed serum proteins were identified. The enrichment and conventional methods yielded similar numbers of total (315 vs. 309), exclusive (17 vs. 11), and major bone-related proteins (18 vs. 14). Most of the adsorbed proteins (n = 298) were common to both extraction groups and included several growth factors, extracellular matrix (ECM) proteins, cell adhesion molecules, and angiogenesis mediators involved in bone regeneration. Functional analyses revealed significant enrichment of ECM, exosomes, immune response, and cell growth components. Key proteins [transforming growth factor-beta 1 (TGFβ1), insulin-like growth factor binding proteins (IGFBP-5, -6, -7)] were exclusively detected with the enrichment-based method. In summary, native collagen membranes exhibited a high protein adsorption capacity in vitro. While both extraction methods were effective, the enrichment-based method showed distinct advantages in detecting specific bone-related proteins. Therefore, the use of multiple extraction methods is advisable in studies investigating protein adsorption on biomaterials.
Keywords: collagen membranes; guided bone regeneration; guided tissue regeneration; mass spectrometry; protein extraction.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
-
- Wang J., Chen X., Guo B., Yang X., Zhou Y., Zhu X., Zhang K., Fan Y., Tu C., Zhang X. A serum protein adsorption profile on BCP ceramics and influence of the elevated adsorption of adhesive proteins on the behaviour of MSCs. J. Mater. Chem. B. 2018;6:7383–7395. doi: 10.1039/C8TB02283F. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

