Genetic and Evolutionary Analysis of Canine Coronavirus in Guangxi Province, China, for 2021-2024
- PMID: 39453048
- PMCID: PMC11512276
- DOI: 10.3390/vetsci11100456
Genetic and Evolutionary Analysis of Canine Coronavirus in Guangxi Province, China, for 2021-2024
Abstract
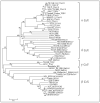
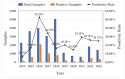
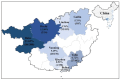
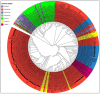
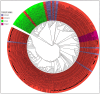
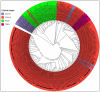
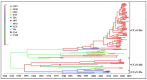
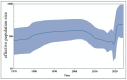
Canine coronavirus (CCoV) is an important gastrointestinal pathogen that causes serious harm to pet dogs worldwide. Here, 1791 clinical samples were collected from pet dogs in different pet hospitals in Guangxi Province, southern China, for the 2021-2024 period and detected for CCoV by a multiplex RT-qPCR. The results showed that 8.43% (151/1791) of samples were positive for CCoV. Sixty-five positive samples were selected to amplify, sequence, and analyze S, M, and N genes. A sequence comparison revealed that the nucleotide and amino acid similarities of the S, M, and N genes were 94.86% and 94.62%, 96.85% and 97.80%, and 96.85% and 97.80%, respectively. Phylogenetic analysis indicated that 65 CCoV strains obtained in this study belonged to the CCoV-II genotype, of which 56 CCoV strains belonged to the CCoV-IIa subtype and 9 CCoV strains belonged to the CCoV-IIb subtype. A potential recombination event analysis of S gene sequences indicated that two CCoV strains, i.e., GXBSHM0328-34 and GXYLAC0318-35, have recombination signals. A Bayesian analysis indicated that the evolutionary rates of the S, M, and N genes were 1.791 × 10-3, 6.529 × 10-4, and 4.775 × 10-4 substitutions/site/year, respectively. The population size grew slowly before 1980 and then began to shrink slowly; it then shrank rapidly in 2005 and expanded sharply in 2020, leveling off thereafter. These results indicated the CCoV strains prevalent in Guangxi Province, southern China, showed a high level of genetic diversity and maintained continuous variation among clinical epidemic strains.
Keywords: M gene; N gene; S gene; canine coronavirus (CCoV); genetic diversity; phylogenetic analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Co-Circulation of Canine Coronavirus I and IIa/b with High Prevalence and Genetic Diversity in Heilongjiang Province, Northeast China.PLoS One. 2016 Jan 15;11(1):e0146975. doi: 10.1371/journal.pone.0146975. eCollection 2016. PLoS One. 2016. PMID: 26771312 Free PMC article.
-
Genetic and Phylogenetic Analysis of Feline Coronavirus in Guangxi Province of China from 2021 to 2024.Vet Sci. 2024 Sep 25;11(10):455. doi: 10.3390/vetsci11100455. Vet Sci. 2024. PMID: 39453047 Free PMC article.
-
Etiology and genetic evolution of canine coronavirus circulating in five provinces of China, during 2018-2019.Microb Pathog. 2020 Aug;145:104209. doi: 10.1016/j.micpath.2020.104209. Epub 2020 Apr 18. Microb Pathog. 2020. PMID: 32311431 Free PMC article.
-
Epidemiological investigation of canine coronavirus infection in Chinese domestic dogs: A systematic review and data synthesis.Prev Vet Med. 2022 Dec;209:105792. doi: 10.1016/j.prevetmed.2022.105792. Epub 2022 Oct 26. Prev Vet Med. 2022. PMID: 36327776
-
A One Health Perspective on Canine Coronavirus: A Wolf in Sheep's Clothing?Microorganisms. 2023 Apr 2;11(4):921. doi: 10.3390/microorganisms11040921. Microorganisms. 2023. PMID: 37110344 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

