Optimizing Scorpion Toxin Processing through Artificial Intelligence
- PMID: 39453213
- PMCID: PMC11511117
- DOI: 10.3390/toxins16100437
Optimizing Scorpion Toxin Processing through Artificial Intelligence
Abstract
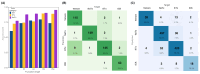
Scorpion toxins are relatively short cyclic peptides (<150 amino acids) that can disrupt the opening/closing mechanisms in cell ion channels. These peptides are widely studied for several reasons including their use in drug discovery. Although improvements in RNAseq have greatly expedited the discovery of new scorpion toxins, their annotation remains challenging, mainly due to their small size. Here, we present a new pipeline to annotate toxins from scorpion transcriptomes using a neural network approach. This pipeline implements basic neural networks to sort amino acid sequences to find those that are likely toxins and thereafter predict the type of toxin represented by the sequence. We anticipate that this pipeline will accelerate the classification of scorpion toxins in forthcoming scorpion genome sequencing projects and potentially serve a useful role in identifying targets for drug development.
Keywords: RNAseq; neural network; python; sodium channel toxins.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

