Leveraging Climate Data for Dengue Forecasting in Ba Ria Vung Tau Province, Vietnam: An Advanced Machine Learning Approach
- PMID: 39453277
- PMCID: PMC11511084
- DOI: 10.3390/tropicalmed9100250
Leveraging Climate Data for Dengue Forecasting in Ba Ria Vung Tau Province, Vietnam: An Advanced Machine Learning Approach
Erratum in
-
Correction: Tuan, D.A. Leveraging Climate Data for Dengue Forecasting in Ba Ria Vung Tau Province, Vietnam: An Advanced Machine Learning Approach. Trop. Med. Infect. Dis. 2024, 9, 250.Trop Med Infect Dis. 2025 May 21;10(5):144. doi: 10.3390/tropicalmed10050144. Trop Med Infect Dis. 2025. PMID: 40423375 Free PMC article.
Abstract
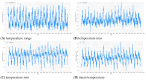
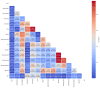
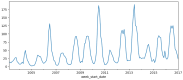
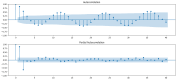
Dengue fever is a persistent public health issue in tropical regions, including Vietnam, where climate variability plays a crucial role in disease transmission dynamics. This study focuses on developing climate-based machine learning models to forecast dengue outbreaks in Ba Ria Vung Tau (BRVT) province, Vietnam, using meteorological data from 2003 to 2022. We utilized four predictive models-Negative Binomial Regression (NBR), Seasonal AutoRegressive Integrated Moving Average with Exogenous Regressors (SARIMAX), Extreme Gradient Boosting (XGBoost) v2.0.3, and long short-term memory (LSTM)-to predict weekly dengue incidence. Key climate variables, including temperature, humidity, precipitation, and wind speed, were integrated into these models, with lagged variables included to capture delayed climatic effects on dengue transmission. The NBR model demonstrated the best performance in terms of predictive accuracy, achieving the lowest Mean Absolute Error (MAE), compared to other models. The inclusion of lagged climate variables significantly enhanced the model's ability to predict dengue cases. Although effective in capturing seasonal trends, SARIMAX and LSTM models struggled with overfitting and failed to accurately predict short-term outbreaks. XGBoost exhibited moderate predictive power but was sensitive to overfitting, particularly without fine-tuning. Our findings confirm that climate-based machine learning models, particularly the NBR model, offer valuable tools for forecasting dengue outbreaks in BRVT. However, improving the models' ability to predict short-term peaks remains a challenge. The integration of meteorological data into early warning systems is crucial for public health authorities to plan timely and effective interventions. This research contributes to the growing body of literature on climate-based disease forecasting and underscores the need for further model refinement to address the complexities of dengue transmission in highly endemic regions.
Keywords: Ba Ria Vung Tau; LSTM; SARIMAX; Vietnam; XGBoost; climate forecasting; dengue fever; machine learning; negative binomial regression.
Conflict of interest statement
The author declares no conflicts of interest.
Figures












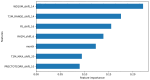
References
-
- Chen J., Ding R.-L., Liu K.-K., Xiao H., Hu G., Xiao X., Yue Q., Lu J.-H., Han Y., Bu J. Collaboration between meteorology and public health: Predicting the dengue epidemic in Guangzhou, China, by meteorological parameters. Front. Cell. Infect. Microbiol. 2022;12:881745. doi: 10.3389/fcimb.2022.881745. - DOI - PMC - PubMed
-
- Colón-González F.J., Soares Bastos L., Hofmann B., Hopkin A., Harpham Q., Crocker T., Amato R., Ferrario I., Moschini F., James S. Probabilistic seasonal dengue forecasting in Vietnam: A modelling study using superensembles. PLoS Med. 2021;18:e1003542. doi: 10.1371/journal.pmed.1003542. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

