Inhibiting lncRNA NEAT1 Increases Glioblastoma Response to TMZ by Reducing Connexin 43 Expression
- PMID: 39453684
- PMCID: PMC11505515
- DOI: 10.1002/cnr2.70031
Inhibiting lncRNA NEAT1 Increases Glioblastoma Response to TMZ by Reducing Connexin 43 Expression
Abstract
Objectives: Glioblastoma multiforme (GBM) is considered the most assailant subtype of gliomas, presenting a formidable obstacle because of its inherent resistance to temozolomide (TMZ). This study aimed to characterize the function of lncRNA NEAT1 in facilitating the advancement of gliomas.
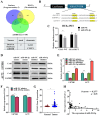
Methods: The expression level of NEAT1 in glioma tissues and cells was detected by qRT-PCR. RNA interference experiment, cell proliferation assay, FITC/PI detection assay, immunoblotting, bioinformatics prediction, a double luciferase reporter gene assay, RNA immunoprecipitation (RIP) assay, SLDT assay and correlation analysis of clinical samples were performed to explore the regulatory effects of NEAT1, miR-454-3p and Cx43 and their role in malignant progression of GBM. The role of NEAT1 in vivo was investigated by an intracranial tumor formation experiment in mice.
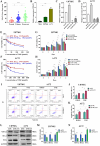
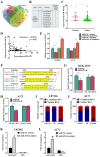
Results: The results showed that recurring gliomas displayed elevated levels of NEAT1 compared to primary gliomas. The suppression of NEAT1 led to a restoration of sensitivity in GBM cells to TMZ. NEAT1 functioned as a competitive endogenous RNA against miR-454-3p. Connexin 43 was identified as a miR-454-3p target. NEAT1 was found to regulate gap junctional intercellular communication by modulating Connexin 43, thereby impacting the response of GBM cells to TMZ chemotherapy. Downregulation of NEAT1 resulted in enhanced chemosensitivity to TMZ and extended the survival of mice.
Conclusions: Overall, these results indicated that the NEAT1/miR-454-3p/Connexin 43 pathway influences GBM cell response to TMZ and could offer a potential new strategy for treating GBM.
Keywords: Connexin 43; NEAT1; chemotherapy sensitivity; miR‐454‐3p; temozolomide.
© 2024 The Author(s). Cancer Reports published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Kanderi T. and Gupta V., Glioblastoma Multiforme (Treasure Island, FL: StatPearls, 2024). - PubMed
MeSH terms
Substances
Grants and funding
- 2021GXNSFBA220024/Natural Science Foundation of Guangxi Province
- GUIKE AD22035055/Specific Research Project of Guangxi for Research Bases and Talents
- 202140240/The Clinical Research Project of Shanghai Municipal Health Commission
- GZxk-z-20-75/Guangxi traditional Chinese medicine key discipline construction project
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

