Arabidopsis uses a molecular grounding mechanism and a biophysical circuit breaker to limit floral abscission signaling
- PMID: 39453742
- PMCID: PMC11536089
- DOI: 10.1073/pnas.2405806121
Arabidopsis uses a molecular grounding mechanism and a biophysical circuit breaker to limit floral abscission signaling
Abstract
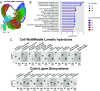
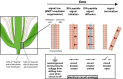
Abscission is the programmed separation of plant organs. It is widespread in the plant kingdom with important functions in development and environmental response. In Arabidopsis, abscission of floral organs (sepals, petals, and stamens) is controlled by two receptor-like protein kinases HAESA (HAE) and HAESA LIKE-2 (HSL2), which orchestrate the programmed dissolution of the abscission zone connecting floral organs to the developing fruit. In this work, we use single-cell RNA sequencing to characterize the core HAE/HSL2 abscission gene expression program. We identify the MAP KINASE PHOSPHATASE-1/MKP1 gene as a negative regulator of this pathway. MKP1 acts prior to activation of HAE/HSL2 signaling to establish a signaling threshold required for the initiation of abscission. Furthermore, we use single-cell data to identify genes expressed in two subpopulations of abscission zone cells: those proximal and those distal to the plane of separation. We identify INFLORESCENCE DEFICIENT IN ABSCISSION/IDA family genes, encoding activating ligands of HAE/HSL2, as enriched in distal abscission zone cells at the base of the abscising organs. We show how this expression pattern forms a biophysical circuit breaker whereby, when the organ is shed, the source of the IDA peptides is removed, leading to cessation of HAE/HSL2 signaling. Overall, this work provides insight into the multiple control mechanisms acting on the abscission-signaling pathway.
Keywords: abscission; signaling; singe-cell rna-sequencing.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures




References
-
- Addicott F. T., Lynch R. S., Physiology of abscission. Ann. Rev. Plant Physiol. 6, 211–238 (1955).
-
- Taylor J. E., Whitelaw C. A., Signals in abscission. New Phytol. 151, 323–340 (2001).
-
- Patharkar O. R., Walker J. C., Connections between abscission, dehiscence, pathogen defense, drought tolerance, and senescence. Plant Sci. 284, 25–29 (2019). - PubMed
-
- Yu Y., Kellogg E. A., Inflorescence abscission zones in grasses: Diversity and genetic regulation. Ann. Plant Rev. 1, 1–35 (2018).
-
- Dutta S. K., Gurung G., Yadav A., Laha R., Mishra V. K., Factors associated with citrus fruit abscission and management strategies developed so far: A review. N Zeal. J. Crop Horticultural Sci. 51, 467–488 (2022).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

