Protocol for detecting genomic insulators in Drosophila using insulator-seq, a massively parallel reporter assay
- PMID: 39453817
- PMCID: PMC11541826
- DOI: 10.1016/j.xpro.2024.103391
Protocol for detecting genomic insulators in Drosophila using insulator-seq, a massively parallel reporter assay
Abstract
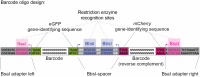
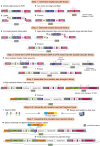
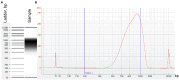
Genomic insulators are DNA elements that prevent transcriptional activation of a promoter by an enhancer when interposed. We present a protocol for insulator-seq that enables high-throughput screening of genomic insulators using a plasmid-based massively parallel reporter assay in Drosophila cultured cells. We describe steps for insulator reporter plasmid library generation, transient transfection into cultured cells, and sequencing library preparation and provide a pipeline for data analysis. For complete details on the use and execution of this protocol, please refer to Tonelli et al.1.
Keywords: Gene Expression; Model Organisms; RNA-seq; Sequencing.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Tonelli A., Cousin P., Jankowski A., Wang B., Dorier J., Barraud J., Zunjarrao S., Gambetta M.C. Systematic screening of enhancer-blocking insulators in Drosophila identifies their DNA sequence determinants. Dev. Cell. 2024;60 doi: 10.1016/j.devcel.2024.10.017. - DOI
-
- Kaushal A., Mohana G., Dorier J., Özdemir I., Omer A., Cousin P., Semenova A., Taschner M., Dergai O., Marzetta F., et al. CTCF loss has limited effects on global genome architecture in Drosophila despite critical regulatory functions. Nat. Commun. 2021;12:1011. doi: 10.1038/s41467-021-21366-2. - DOI - PMC - PubMed
-
- Custom code used for the analysis in Tonelli et al., Version v1.0 (Zenodo) 2024. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

