Structural basis for regulation of a CBASS-CRISPR-Cas defense island by a transmembrane anti-σ factor and its ECF σ partner
- PMID: 39454004
- PMCID: PMC11506125
- DOI: 10.1126/sciadv.adp1053
Structural basis for regulation of a CBASS-CRISPR-Cas defense island by a transmembrane anti-σ factor and its ECF σ partner
Abstract
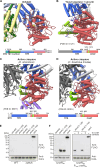
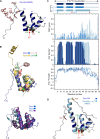
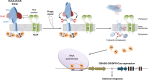
How CRISPR-Cas and cyclic oligonucleotide-based antiphage signaling systems (CBASS) are coordinately deployed against invaders remains unclear. We show that a locus containing two CBASS and one type III-B CRISPR-Cas system, regulated by the transmembrane anti-σ DdvA and its cognate extracytoplasmic function (ECF) σ DdvS, can defend Myxococcus xanthus against a phage. Cryo-electron microscopy reveals DdvA-DdvS pairs assemble as arrow-shaped transmembrane dimers. Each DdvA periplasmic domain adopts a separase/craspase-type tetratricopeptide repeat (TPR)-caspase HetF-associated with TPR (TPR-CHAT) architecture with an incomplete His-Cys active site, lacking three α-helices conserved among CHAT domains. Each active site faces the dimer interface, raising the possibility that signal-induced caspase-like DdvA autoproteolysis in trans precedes RseP-mediated intramembrane proteolysis and DdvS release. Nuclear magnetic resonance reveals a DdvA cytoplasmic CHCC-type zinc-bound three-helix bundle that binds to DdvS σ2 and σ4 domains, undergoing σ4-induced helix extension to trap DdvS. Altogether, we provide structural-mechanistic insights into membrane anti-σ-ECF σ regulation of an antiviral CBASS-CRISPR-Cas defense island.
Figures





References
-
- Millman A., Melamed S., Leavitt A., Doron S., Bernheim A., Hör J., Garb J., Bechon N., Brandis A., Lopatina A., Ofir G., Hochhauser D., Stokar-Avihail A., Tal N., Sharir S., Voichek M., Erez Z., Ferrer J. L. M., Dar D., Kacen A., Amitai G., Sorek R., An expanded arsenal of immune systems that protect bacteria from phages. Cell Host Microbe 30, 1556–1569.e5 (2022). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

