Stepwise virus assembly in the cell nucleus revealed by spatiotemporal click chemistry of DNA replication
- PMID: 39454009
- PMCID: PMC11506174
- DOI: 10.1126/sciadv.adq7483
Stepwise virus assembly in the cell nucleus revealed by spatiotemporal click chemistry of DNA replication
Abstract
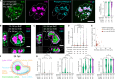
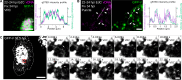
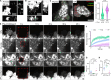
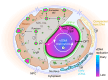
Biomolecular assemblies are fundamental to life and viral disease. The spatiotemporal coordination of viral replication and assembly is largely unknown. Here, we developed a dual-color click chemistry procedure for imaging adenovirus DNA (vDNA) replication in the cell nucleus. Late- but not early-replicated vDNA was packaged into virions. Early-replicated vDNA segregated from the viral replication compartment (VRC). Single object tracking, superresolution microscopy, fluorescence recovery after photobleaching, and correlative light-electron microscopy revealed a stepwise assembly program involving vDNA and capsid intermediates. Depending on replication and the scaffolding protein 52K, late-replicated vDNA with rapidly exchanging green fluorescent protein-tagged capsid linchpin protein V and incomplete virions emerged from the VRC periphery. These nanogel-like puncta exhibited restricted movements and were located with the capsid proteins hexon, VI, and virions in the nuclear periphery, suggestive of sites for virion formation. Our findings identify VRC dynamics and assembly intermediates, essential for stepwise productive adenovirus morphogenesis.
Figures




References
-
- Trinkle-Mulcahy L., Lamond A. I., Toward a high-resolution view of nuclear dynamics. Science 318, 1402–1407 (2007). - PubMed
-
- Mitchison T. J., Salmon E. D., Mitosis: A history of division. Nat. Cell Biol. 3, E17–E21 (2001). - PubMed
-
- Mei Y., Liu Y., Ji P., Understanding terminal erythropoiesis: An update on chromatin condensation, enucleation, and reticulocyte maturation. Blood Rev. 46, 100740 (2021). - PubMed
-
- Hao S. L., Ni F. D., Yang W. X., The dynamics and regulation of chromatin remodeling during spermiogenesis. Gene 706, 201–210 (2019). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

