TopoTxR: A topology-guided deep convolutional network for breast parenchyma learning on DCE-MRIs
- PMID: 39454312
- PMCID: PMC12228977
- DOI: 10.1016/j.media.2024.103373
TopoTxR: A topology-guided deep convolutional network for breast parenchyma learning on DCE-MRIs
Abstract
Characterization of breast parenchyma in dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) is a challenging task owing to the complexity of underlying tissue structures. Existing quantitative approaches, like radiomics and deep learning models, lack explicit quantification of intricate and subtle parenchymal structures, including fibroglandular tissue. To address this, we propose a novel topological approach that explicitly extracts multi-scale topological structures to better approximate breast parenchymal structures, and then incorporates these structures into a deep-learning-based prediction model via an attention mechanism. Our topology-informed deep learning model, TopoTxR, leverages topology to provide enhanced insights into tissues critical for disease pathophysiology and treatment response. We empirically validate TopoTxR using the VICTRE phantom breast dataset, showing that the topological structures extracted by our model effectively approximate the breast parenchymal structures. We further demonstrate TopoTxR's efficacy in predicting response to neoadjuvant chemotherapy. Our qualitative and quantitative analyses suggest differential topological behavior of breast tissue in treatment-naïve imaging, in patients who respond favorably to therapy as achieving pathological complete response (pCR) versus those who do not. In a comparative analysis with several baselines on the publicly available I-SPY 1 dataset (N = 161, including 47 patients with pCR and 114 without) and the Rutgers proprietary dataset (N = 120, with 69 patients achieving pCR and 51 not), TopoTxR demonstrates a notable improvement, achieving a 2.6% increase in accuracy and a 4.6% enhancement in AUC compared to the state-of-the-art method.
Keywords: 3D CNN; DCE-MRI; Persistent homology; Spatial attention; Topology; pCR prediction.
Copyright © 2024 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest All authors disclosed no relevant relationships.
Figures

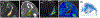

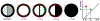
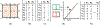



References
-
- Adams H, Emerson T, Kirby M, Neville R, Peterson C, Shipman P, Chepushtanova S, Hanson E, Motta F, Ziegelmeier L, 2017. Persistence images: A stable vector representation of persistent homology. J. Mach .
-
- Akazawa K, Tamaki Y, Taguchi T, Tanji Y, Miyoshi Y, Kim SJ, Ueda S, Yanagisawa T, Sato Y, Tamura S, et al. , 2006. Preoperative evaluation of residual tumor extent by three-dimensional magnetic resonance imaging in breast cancer patients treated with neoadjuvant chemotherapy. The breast journal 12, 130–137. - PubMed
-
- Arasu VA, Miglioretti DL, Sprague BL, Alsheik NH, Buist DS, Henderson LM, Herschorn SD, Lee JM, Onega T, Rauscher GH, et al. , 2019. Population-based assessment of the association between magnetic resonance imaging background parenchymal enhancement and future primary breast cancer risk. Journal of Clinical Oncology 37, 954. - PMC - PubMed
-
- Badano A, Graff CG, Badal A, Sharma D, Zeng R, Samuelson FW, Glick SJ, Myers KJ, 2018. Evaluation of Digital Breast Tomosynthesis as Replacement of Full-Field Digital Mammography Using an In Silico Imaging Trial. JAMA Network Open 1, e185474–e185474. URL: https://doi.org/10.1001/jamanetworkopen.2018.5474, doi: 10.1001/jamanetworkopen.2018.5474. - DOI - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

