Novel risk loci in LGI1-antibody encephalitis: genome-wide association study discovery and validation cohorts
- PMID: 39454566
- PMCID: PMC11884648
- DOI: 10.1093/brain/awae349
Novel risk loci in LGI1-antibody encephalitis: genome-wide association study discovery and validation cohorts
Abstract
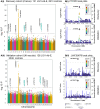
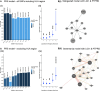
Encephalitis with antibodies to leucine-rich glioma-inactivated 1 (LGI1-Ab-E) is a common form of autoimmune encephalitis, presenting with seizures and neuropsychiatric changes, predominantly in older males. More than 90% of patients carry the human leukocyte antigen (HLA) class II allele, HLA-DRB1*07:01. However, this is also present in 25% of healthy controls. Therefore, we hypothesized the presence of additional genetic predispositions. In this genome-wide association study and meta-analysis, we studied a discovery cohort of 131 French LGI1-Ab-E and a validation cohort of 126 American, British and Irish LGI1-Ab-E patients, ancestry-matched to 2613 and 2538 European controls, respectively. Outside the known major HLA signal, we found two single nucleotide polymorphisms at genome-wide significance (P < 5 × 10-8), implicating PTPRD, a protein tyrosine phosphatase, and LINC00670, a non-protein coding RNA gene. Meta-analysis defined four additional non-HLA loci, including the protein coding COBL gene. Polygenic risk scores with and without HLA variants proposed a contribution of non-HLA loci. In silico network analyses suggested LGI1 and PTPRD-mediated interactions via the established receptors of LGI1, ADAM22 and ADAM23. Our results identify new genetic loci in LGI1-Ab-E. These findings present opportunities for mechanistic studies and offer potential markers of susceptibility, prognostics and therapeutic responses.
Keywords: GWAS; HLA; LGI1; PTPRD; encephalitis; genetics.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
K.S.E. is now an employee of Arcturis Data. A.R.H. is a current employee and/or stockholder of AstraZeneca. A.E.H. has received funding from UCB-Pharma. N. M. has received honoraria for lecturing and travel expenses for attending meetings from Biogen Idec, GlaxoSmith Kline, Teva, Novartis Pharma, Bayer Healthcare, Genzyme, Alexion Pharmaceuticals, Fresenius Medical Care, Diamed, UCB Pharma, AngeliniPharma, BIAL and Sanofi-Aventis, has received royalties for consulting from UCB Pharma, Alexion Pharmaceuticals and Sanofi-Aventis and has received financial research support from Euroimmun, Fresenius Medical Care, Diamed, Alexion Pharmaceuticals and Novartis Pharma. F.L. discloses speaker honoraria from Grifols, Teva, Biogen, Bayer, Roche, Novartis, Fresenius, travel funding from Merck, Grifols and Bayer and serving on advisory boards for Roche, Biogen and Alexion. S.R.I. has received honoraria/research support from UCB, Immunovant, MedImmun, Roche, Janssen, Cerebral therapeutics, ADC therapeutics, Brain, CSL Behring and ONO Pharma; licensed royalties on patent application WO/2010/046716 entitled ‘Neurological Autoimmune Disorders’ and has filed two other patents entitled ‘Diagnostic method and therapy’ (WO2019211633 and US-2021-0071249-A1; PCT application WO202189788A1) and ‘Biomarkers’ (PCT/GB2022/050614 and WO202189788A1). The remaining authors report no competing interests.
Figures
References
-
- Zuliani L, Marangoni S, De Gaspari P, et al. Epidemiology of neuronal surface antibody-mediated autoimmune encephalitis and antibody-based diagnostics. J Neuroimmunol. 2021;357:577598. - PubMed
-
- Kim TJ, Lee ST, Moon J, et al. Anti-LGI1 encephalitis is associated with unique HLA subtypes. Ann Neurol. 2017;81:183–192. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

