Sugar transporter modulates nitrogen-determined tillering and yield formation in rice
- PMID: 39455567
- PMCID: PMC11512014
- DOI: 10.1038/s41467-024-53651-1
Sugar transporter modulates nitrogen-determined tillering and yield formation in rice
Abstract
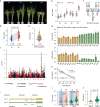
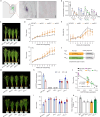
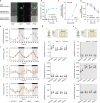
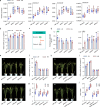
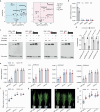
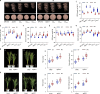
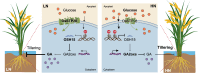
Nitrogen (N) fertilizer application ensures crop production and food security worldwide. N-controlled boosting of shoot branching that is also referred as tillering can improve planting density for increasing grain yield of cereals. Here, we report that Sugar Transporter Protein 28 (OsSTP28) as a key regulator of N-responsive tillering and yield formation in rice. N supply inhibits the expression of OsSTP28, resulting in glucose accumulation in the apoplast of tiller buds, which in turn suppresses the expression of a transcriptional inhibitor ORYZA SATIVA HOMEOBOX 15 (OSH15) via an epigenetic mechanism to activate gibberellin 2-oxidases (GA2oxs)-facilitated gibberellin catabolism in shoot base. Thereby, OsSTP28-OSH15-GA2oxs module reduces the level of bioactive gibberellin in shoot base upon increased N supply, and consequently promotes tillering and grain yield. Moreover, we identify an elite allele of OsSTP28 that can effectively promote N-responsive tillering and yield formation, thus representing a valuable breeding target of N use efficiency improvement for agricultural sustainability.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Research Materials

