Biallelic GGGCC repeat expansion leading to NAXE-related mitochondrial encephalopathy
- PMID: 39455596
- PMCID: PMC11512015
- DOI: 10.1038/s41525-024-00429-5
Biallelic GGGCC repeat expansion leading to NAXE-related mitochondrial encephalopathy
Abstract
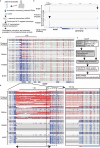
Repeat expansions cause at least 50 hereditary disorders, including Friedreich ataxia and other diseases known to cause mitochondrial dysfunction. We identified a patient with NAXE-related mitochondrial encephalopathy and novel biallelic GGGCC repeat expansion as long as ~200 repeats in the NAXE promoter region using long-read sequencing. In addition to a marked reduction in the RNA and protein, we found a marked reduction in nascent RNA in the promoter using native elongating transcript-cap analysis of gene expression (NET-CAGE), suggesting transcriptional suppression. Accordingly, CpG hypermethylation was observed in the repeat region. Genetic analyses determined that homozygosity in the patient was due to maternal chromosome 1 uniparental disomy (UPD). We assessed short variants within NAXE including the repeat region in the undiagnosed mitochondrial encephalopathy cohort of 242 patients. This study identified the GGGCC repeat expansion causing a mitochondrial disease and suggests that UPD could significantly contribute to homozygosity for rare repeat-expanded alleles.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


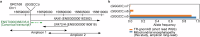
References
Grants and funding
- JP23ek0109625/Japan Agency for Medical Research and Development (AMED)
- JP23ek0109672/Japan Agency for Medical Research and Development (AMED)
- JP23kk0305024/Japan Agency for Medical Research and Development (AMED)
- JP23zf0127001/Japan Agency for Medical Research and Development (AMED)
- JP22ek0109485/Japan Agency for Medical Research and Development (AMED)
LinkOut - more resources
Full Text Sources
Miscellaneous

