Development of a high-throughput screening system targeting the protein-protein interactions between PRL and CNNM
- PMID: 39455715
- PMCID: PMC11511866
- DOI: 10.1038/s41598-024-76269-1
Development of a high-throughput screening system targeting the protein-protein interactions between PRL and CNNM
Abstract
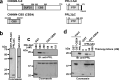
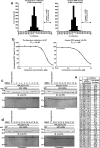
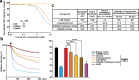
Phosphatase of regenerating liver (PRL) is an oncogenic protein that promotes tumor progression by directly binding to cyclin M (CNNM) membrane proteins and inhibiting their Mg2+ efflux activity. In this study, we have developed a high-throughput screening system to detect the interactions between PRL and CNNM proteins based on homogenous time-resolved fluorescence resonance energy transfer (HTR-FRET, HTRF). We optimized the tag sequences attached to the recombinant proteins of the CNNM4 CBS domains and PRL3 lacking the carboxyl terminal CAAX motif, and successfully detected the interaction by observing the FRET signal in the mixture of the tagged proteins and fluorophore-conjugated antibodies. Moreover, we performed compound library screening using this system and discovered several compounds that could efficiently inhibit the PRL-CNNM interaction. Characterization of one candidate compound revealed that it was relatively stable compared with thienopyridone, a known inhibitor of the PRL-CNNM interaction. The candidate compound can also inhibit PRL function in cells: suppression of CNNM-dependent Mg2+ efflux, and has sufficient in vitro drug metabolism and pharmacokinetic properties. Overall, these results demonstrate the effectiveness of this screening system for identifying novel inhibitors of the PRL-CNNM interaction, which could contribute to the development of novel anti-cancer drugs.
Keywords: Compound screening; Cyclin M (CNNM); HTRF); Homogenous time-resolved fluorescence resonance energy transfer (HTR-FRET; Phosphatase of regenerating liver (PRL); Protein-protein interaction (PPI).
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Saha, S. et al. A phosphatase associated with metastasis of colorectal cancer. Science 294, 1343–1346 (2001). - PubMed
-
- Bessette, D. C., Qiu, D. & Pallen, C. J. PRL PTPs: mediators and markers of cancer progression. Cancer Metastasis Rev. 27, 231–252 (2008). - PubMed
-
- Al-Aidaroos, A. Q. & Zeng, Q. PRL-3 phosphatase and cancer metastasis. J. Cell. Biochem. 111, 1087–1098 (2010). - PubMed
-
- Kozlov, G. et al. Structural insights into molecular function of the metastasis-associated phosphatase PRL-3. J. Biol. Chem. 279, 11882–11889 (2004). - PubMed

