ALYREF recruits ELAVL1 to promote colorectal tumorigenesis via facilitating RNA m5C recognition and nuclear export
- PMID: 39455812
- PMCID: PMC11512073
- DOI: 10.1038/s41698-024-00737-0
ALYREF recruits ELAVL1 to promote colorectal tumorigenesis via facilitating RNA m5C recognition and nuclear export
Abstract
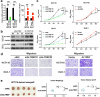
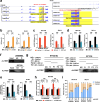
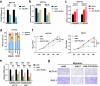
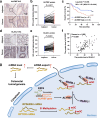
ALYREF can recognize 5-methylcytosine (m5C) decoration throughout RNAs to regulate RNA metabolism. However, its implications in cancer and precise regulatory mechanisms remain largely elusive. Here, we demonstrated that ALYREF supported colorectal cancer (CRC) growth and migration. Integrated analysis of ALYREF-RIP-Bis-seq and transcriptome profiles identified ribosomal protein S6 kinase B2 (RPS6KB2) and regulatory-associated protein of mTOR (RPTOR) as ALYREF's possible downstream effectors. Mechanistically, ALYREF formed a complex with ELAV like RNA binding protein 1 (ELAVL1) to cooperatively promote m5C recognition and nuclear export of the two mRNAs. Moreover, ALYREF protein was highly expressed in tumor tissues of CRC patients, which predicted their poor prognosis. E2F transcription factor 6 (E2F6)-mediated transactivation gave a molecular insight into ALYREF overexpression. Collectively, ALYREF recruits ELAVL1 to collaboratively facilitate m5C recognition and nuclear export of RPS6KB2 and RPTOR transcripts for colorectal tumorigenesis, providing RNA m5C methylation as promising therapeutic targets and prognostic biomarkers for CRC.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
ALYREF-mediated RNA 5-Methylcytosine modification Promotes Hepatocellular Carcinoma Progression Via Stabilizing EGFR mRNA and pSTAT3 activation.Int J Biol Sci. 2024 Jan 1;20(1):331-346. doi: 10.7150/ijbs.82316. eCollection 2024. Int J Biol Sci. 2024. PMID: 38164181 Free PMC article.
-
ALYREF m5C RNA methylation reader predicts bladder cancer prognosis by regulating the tumor immune microenvironment.Medicine (Baltimore). 2024 Apr 5;103(14):e37590. doi: 10.1097/MD.0000000000037590. Medicine (Baltimore). 2024. PMID: 38579085 Free PMC article.
-
The role of the HIF-1α/ALYREF/PKM2 axis in glycolysis and tumorigenesis of bladder cancer.Cancer Commun (Lond). 2021 Jul;41(7):560-575. doi: 10.1002/cac2.12158. Epub 2021 May 15. Cancer Commun (Lond). 2021. PMID: 33991457 Free PMC article.
-
ALYREF (Aly/REF export factor): A potential biomarker for predicting cancer occurrence and therapeutic efficacy.Life Sci. 2024 Feb 1;338:122372. doi: 10.1016/j.lfs.2023.122372. Epub 2023 Dec 21. Life Sci. 2024. PMID: 38135116 Review.
-
Biological functions of 5-methylcytosine RNA-binding proteins and their potential mechanisms in human cancers.Front Oncol. 2025 Feb 7;15:1534948. doi: 10.3389/fonc.2025.1534948. eCollection 2025. Front Oncol. 2025. PMID: 39990690 Free PMC article. Review.
Cited by
-
5-methylcytosine regulated CCNL2 promotes tumorigenesis and cisplatin resistance of ovarian cancer with therapeutic implications.J Ovarian Res. 2025 Jul 24;18(1):162. doi: 10.1186/s13048-025-01753-9. J Ovarian Res. 2025. PMID: 40707993 Free PMC article.
-
Exploring the impact of cuproptosis on prostate cancer prognosis via RNA methylation regulation based on single cell and bulk RNA sequencing data.Front Pharmacol. 2025 Apr 1;16:1573611. doi: 10.3389/fphar.2025.1573611. eCollection 2025. Front Pharmacol. 2025. PMID: 40235543 Free PMC article.
-
m5C RNA methylation in cancer: from biological mechanism to clinical perspectives.Eur J Med Res. 2025 Jun 21;30(1):503. doi: 10.1186/s40001-025-02812-z. Eur J Med Res. 2025. PMID: 40544318 Free PMC article. Review.
-
Phosphofructokinase-1 redefined: a metabolic hub orchestrating cancer hallmarks through multi-dimensional control networks.J Transl Med. 2025 Aug 6;23(1):873. doi: 10.1186/s12967-025-06897-2. J Transl Med. 2025. PMID: 40770805 Free PMC article. Review.
References
-
- Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). - PubMed
-
- Siegel, R. L. et al. Global patterns and trends in colorectal cancer incidence in young adults. Gut68, 2179–2185 (2019). - PubMed
-
- Kehm, R. D. et al. Age-specific trends in colorectal cancer incidence for women and men, 1935–2017. Gastroenterology161, 1060–1062.e1063 (2021). - PubMed
-
- Dekker, E., Tanis, P. J., Vleugels, J. L. A., Kasi, P. M. & Wallace, M. B. Colorectal cancer. Lancet394, 1467–1480 (2019). - PubMed
-
- Han, X., Wang, M., Zhao, Y. L., Yang, Y. & Yang, Y. G. RNA methylations in human cancers. Semin. Cancer Biol.75, 97–115 (2021). - PubMed
Grants and funding
- 82002496/National Natural Science Foundation of China (National Science Foundation of China)
- 3502Z20244ZD1003/Xiamen Municipal Bureau of Science and Technology (Xiamen Science and Technology Bureau)
- 3502Z20214ZD3005/Xiamen Municipal Bureau of Science and Technology (Xiamen Science and Technology Bureau)
- 3502Z20204003/Xiamen Municipal Bureau of Science and Technology (Xiamen Science and Technology Bureau)
- 2020J05300/Natural Science Foundation of Fujian Province (Fujian Provincial Natural Science Foundation)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

