The two-stage detection-after-segmentation model improves the accuracy of identifying subdiaphragmatic lesions
- PMID: 39455821
- PMCID: PMC11511953
- DOI: 10.1038/s41598-024-76450-6
The two-stage detection-after-segmentation model improves the accuracy of identifying subdiaphragmatic lesions
Abstract
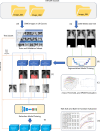
Chest X-rays (CXRs) are primarily used to detect lung lesions. While the abdominal portion of CXRs can sometimes reveal critical conditions, research in this area is limited. To address this, we introduce a two-stage architecture that separates the abdominal region from the CXR and detects abdominal lesions using a specialized dataset. We compared the performance of our method on whole CXRs versus isolated abdominal regions. First, we created masks for the right upper quadrant (RUQ), left upper quadrant (LUQ), and upper abdomen (ABD) regions and trained corresponding segmentation models for each area. For detecting abdominal lesions, we curated a dataset of 5,996 images, categorized into 19 classes based on anatomical locations, air patterns, and levels of stomach or bowel dilation. The detection process was initially conducted on the original images, followed by the three regional areas, RUQ, LUQ, and ABD, extracted by the segmentation models. The results showed that the detection model trained on the entire ABD region achieved the highest accuracy, followed closely by the RUQ and LUQ models. In contrast, the CXR model had the lowest accuracy. This study highlights that the two-stage architecture effectively manages distinct segmentation and detection tasks in CXRs, offering a promising avenue for more accurate diagnoses. It also suggests that an optimal ratio between the sizes of the target lesions and the input images may exist.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
A review on lung boundary detection in chest X-rays.Int J Comput Assist Radiol Surg. 2019 Apr;14(4):563-576. doi: 10.1007/s11548-019-01917-1. Epub 2019 Feb 7. Int J Comput Assist Radiol Surg. 2019. PMID: 30730032 Free PMC article. Review.
-
Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration.IEEE Trans Med Imaging. 2014 Feb;33(2):577-90. doi: 10.1109/TMI.2013.2290491. Epub 2013 Nov 13. IEEE Trans Med Imaging. 2014. PMID: 24239990 Free PMC article.
-
A comprehensive segmentation of chest X-ray improves deep learning-based WHO radiologically confirmed pneumonia diagnosis in children.Eur Radiol. 2024 May;34(5):3471-3482. doi: 10.1007/s00330-023-10367-y. Epub 2023 Nov 6. Eur Radiol. 2024. PMID: 37930411
-
Automatic creation of annotations for chest radiographs based on the positional information extracted from radiographic image reports.Comput Methods Programs Biomed. 2021 Sep;209:106331. doi: 10.1016/j.cmpb.2021.106331. Epub 2021 Aug 4. Comput Methods Programs Biomed. 2021. PMID: 34418813
-
Segmentation and Image Analysis of Abnormal Lungs at CT: Current Approaches, Challenges, and Future Trends.Radiographics. 2015 Jul-Aug;35(4):1056-76. doi: 10.1148/rg.2015140232. Radiographics. 2015. PMID: 26172351 Free PMC article. Review.
References
-
- Pereira, B. M. Abdominal compartment syndrome and intra-abdominal hypertension. Curr. Opin. Crit. Care. 25, 688–696. 10.1097/MCC.0000000000000665 (2019). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

