miR-124 coordinates metabolic regulators acting at early stages of human neurogenesis
- PMID: 39455851
- PMCID: PMC11511827
- DOI: 10.1038/s42003-024-07089-2
miR-124 coordinates metabolic regulators acting at early stages of human neurogenesis
Abstract
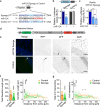
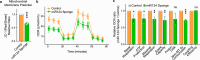
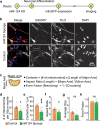
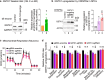
Metabolic dysregulation of neurons is associated with diverse human brain disorders. Metabolic reprogramming occurs during neuronal differentiation, but it is not fully understood which molecules regulate metabolic changes at the early stages of neurogenesis. In this study, we report that miR-124 is a driver of metabolic change at the initiating stage of human neurogenesis. Proteome analysis has shown the oxidative phosphorylation pathway to be the most significantly altered among the differentially expressed proteins (DEPs) in the immature neurons after the knockdown of miR-124. In agreement with these proteomics results, miR-124-depleted neurons display mitochondrial dysfunctions, such as decreased mitochondrial membrane potential and cellular respiration. Moreover, morphological analyses of mitochondria in early differentiated neurons after miR-124 knockdown result in smaller and less mature shapes. Lastly, we show the potential of identified DEPs as novel metabolic regulators in early neuronal development by validating the effects of GSTK1 on cellular respiration. GSTK1, which is upregulated most significantly in miR-124 knockdown neurons, reduces the oxygen consumption rate of neural cells. Collectively, our data highlight the roles of miR-124 in coordinating metabolic maturation at the early stages of neurogenesis and provide insights into potential metabolic regulators associated with human brain disorders characterized by metabolic dysfunctions.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
- R01 AG056306/AG/NIA NIH HHS/United States
- NRF-2022R1A2C3008991/National Research Foundation of Korea (NRF)
- R01 AG056511/AG/NIA NIH HHS/United States
- R01 AG057706/AG/NIA NIH HHS/United States
- RS-2024-00398786/National Research Foundation of Korea (NRF)
- UCSD - R01 AG057706/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- NRF-2022R1C1C2003822/National Research Foundation of Korea (NRF)
- UCSD - R01 AG056511/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- CCSG: P30 014195/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- RS-2024-00335144/National Research Foundation of Korea (NRF)
- NRF-2019R1C1C1010482/National Research Foundation of Korea (NRF)
LinkOut - more resources
Full Text Sources

