Aberrant BCAT1 expression augments MTOR activity and accelerates disease progression in chronic lymphocytic leukemia
- PMID: 39455853
- PMCID: PMC11717693
- DOI: 10.1038/s41375-024-02448-8
Aberrant BCAT1 expression augments MTOR activity and accelerates disease progression in chronic lymphocytic leukemia
Erratum in
-
Correction: Aberrant BCAT1 expression augments MTOR activity and accelerates disease progression in chronic lymphocytic leukemia.Leukemia. 2025 Jan;39(1):274. doi: 10.1038/s41375-024-02472-8. Leukemia. 2025. PMID: 39537979 Free PMC article. No abstract available.
Abstract
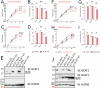
We performed gene expression profiling of mRNA/cDNA isolated from N = 117 flow sorted CLL. We detected aberrant expression of the metabolic enzyme branched chain amino acid transferase (BCAT1) in CLL with del17p/TP53mut. Through extensive validation, we confirmed the highly preferential expression of BCAT1 in CLL with del17p/TP53mut (66%) or trisomy 12 (77%). BCAT1 was not expressed in B cells isolated from normal human lymph nodes. The products of the bidirectional BCAT1 reaction, including leucine, acetyl-CoA, and alpha-ketoglutarate are known activators of MTOR. We measured an ~two-fold higher MTOR activity via normalized p-S6K levels in primary CLL with BCAT1 high versus absent expression before and after sIgM crosslinking. Through steady state metabolomics and heavy isotope metabolic tracing in primary CLL cells, we demonstrate that CLL cells are avid consumers of branched chain amino acids (BCAAs) and that BCAT1 in CLL engages in bidirectional substrate reactions. Of additional interest, CLL with aberrant BCAT1 expression were less sensitive to Venetoclax-induced apoptosis. Biologically, three CLL-derived cell lines with disruption of BCAT1 had substantially reduced growth ex vivo. Clinically, the expression of any detectable BCAT1 protein in CLL independently associated with shorter median survival (125 months versus 296 months; p < 0.0001), even after exclusion of del17p/TP53mut cases.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: Sami Malek owns stock in Abbvie. The other authors declared no conflicts.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

